Fast and accurate short read alignment with BurrowsWheeler

Fast and accurate short read alignment with Burrows–Wheeler transform Heng Li and Richard Durbin∗ Members of this presentation: Yunji Wang Sree Devineni Zhen Gao

�Motivation The first generation of hash table-based methods (e. g. MAQ) are: �Slow �Not support gapped alignment
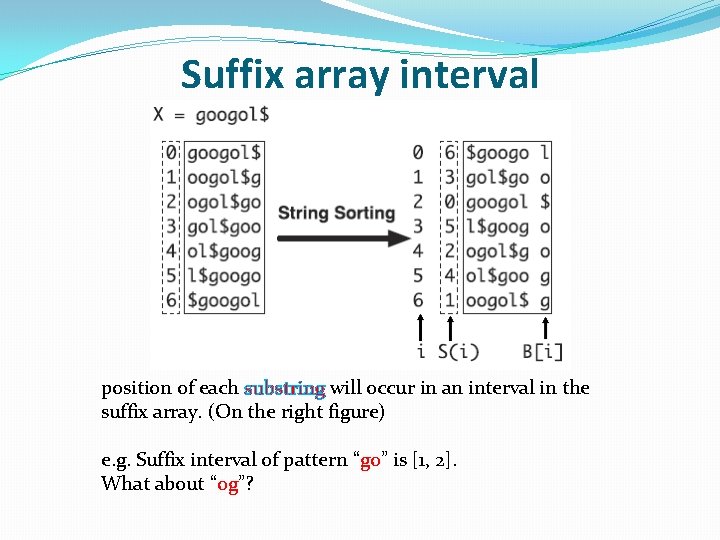
Suffix array interval position of each substring will occur in an interval in the suffix array. (On the right figure) e. g. Suffix interval of pattern “go” is [1, 2]. What about “og”?

Prefix trie and Inexact string matching Prefix trie of string “GOOGOL” The dashed line shows how to find string ‘LOL’ LOL (1 mismatch allowed) What about “LOG”? LOG
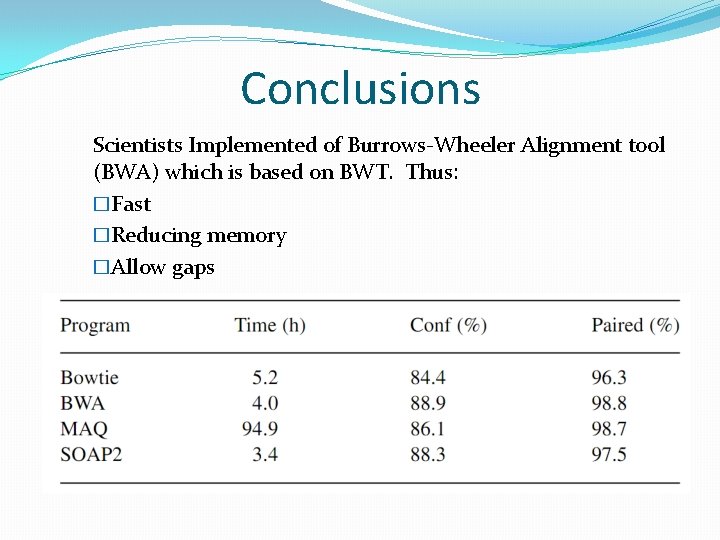
Conclusions Scientists Implemented of Burrows-Wheeler Alignment tool (BWA) which is based on BWT. Thus: �Fast �Reducing memory �Allow gaps

REFERENCES �Heng Li and Richard Durbin (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics, 25, no. 14 2009, pages 1754– 1760
CS 6293: Advanced Topics: Current Bioinformatics A probabilistic framework for aligning paired-end RNA-seq data Members of this presentation: Yunji Wang Sree Devineni Zhen Gao
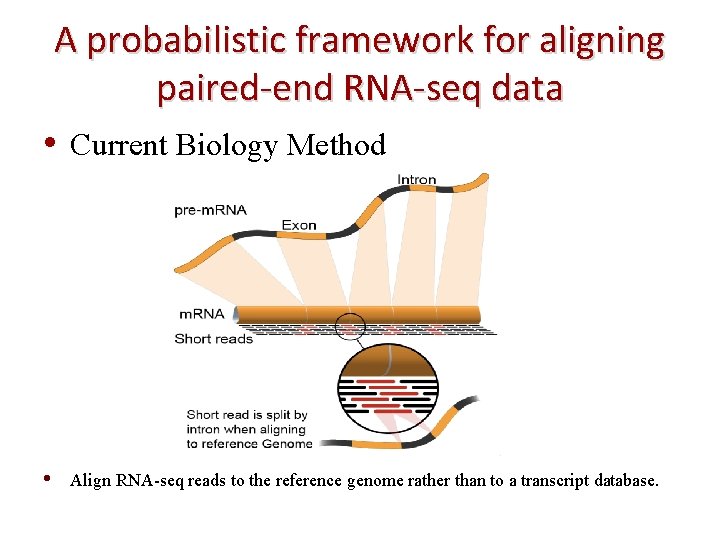
A probabilistic framework for aligning paired-end RNA-seq data • Current Biology Method • Align RNA-seq reads to the reference genome rather than to a transcript database.
Current Biology Problem • • • A single read: Constitute 35 -100 consecutive nucleotides of a fragment of an m. RNA transcript. However, the expected size of m. RNA fragments are around 182 bp. Paired-end read (PER)protocol sequences two ends of a size-selected fragment of an m. RNA. (Double the length of single read)
Problem of PER fragment alignment • Problem: The expected distance between the two end reads within the transcript fragment, know as mate-pair distance The distance between the two ends when aligned to the genome is quit different with mate-pair distance.
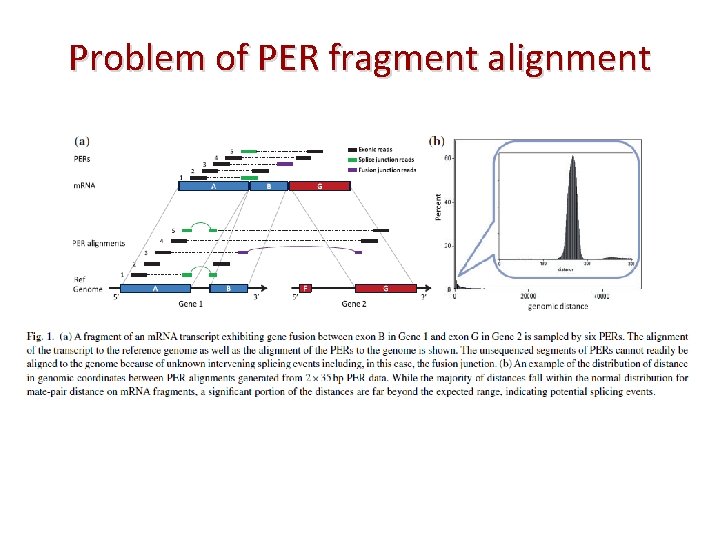
Problem of PER fragment alignment
Current Tools • Top. Hat reports the closest end alignment for a PER. • Splice. Map considers PERs with ends mapped within 400 000 bp on the genome.
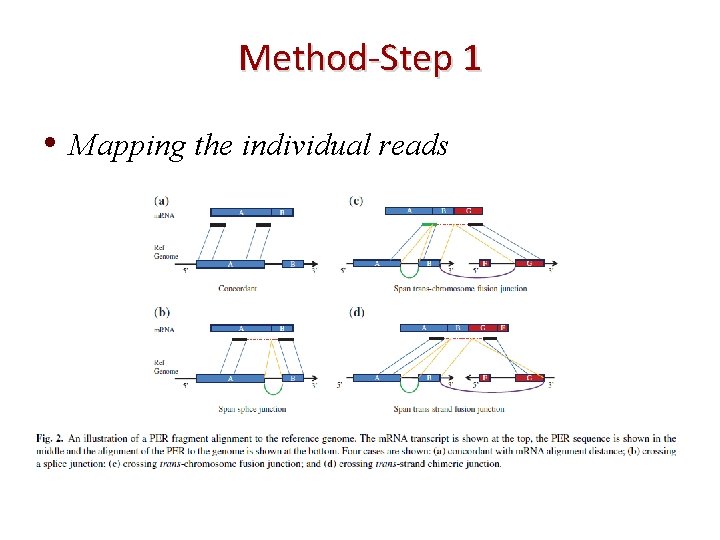
Method-Step 1 • Mapping the individual reads
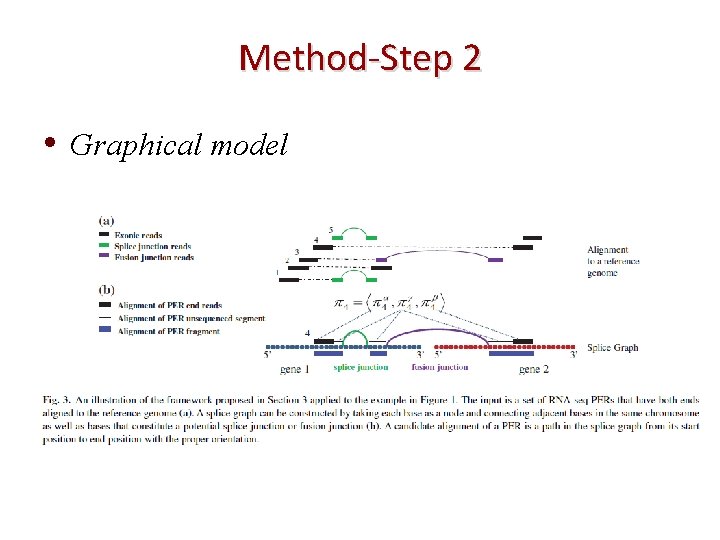
Method-Step 2 • Graphical model
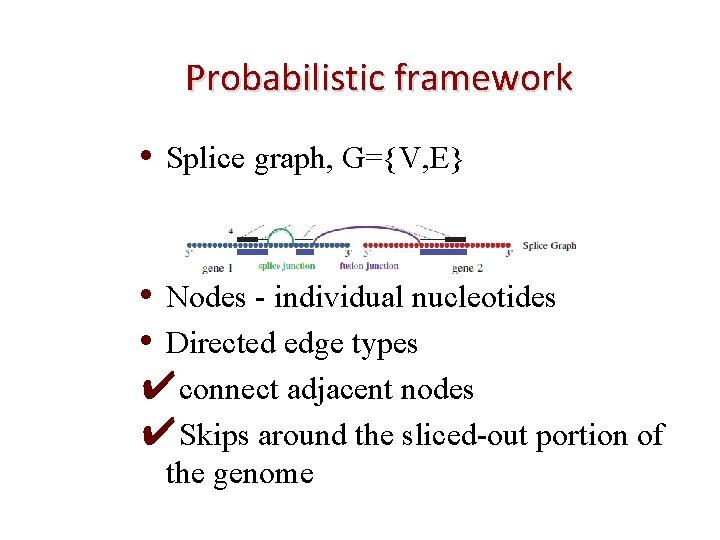
Probabilistic framework • Splice graph, G={V, E} • Nodes - individual nucleotides • Directed edge types ✔connect adjacent nodes ✔Skips around the sliced-out portion of the genome
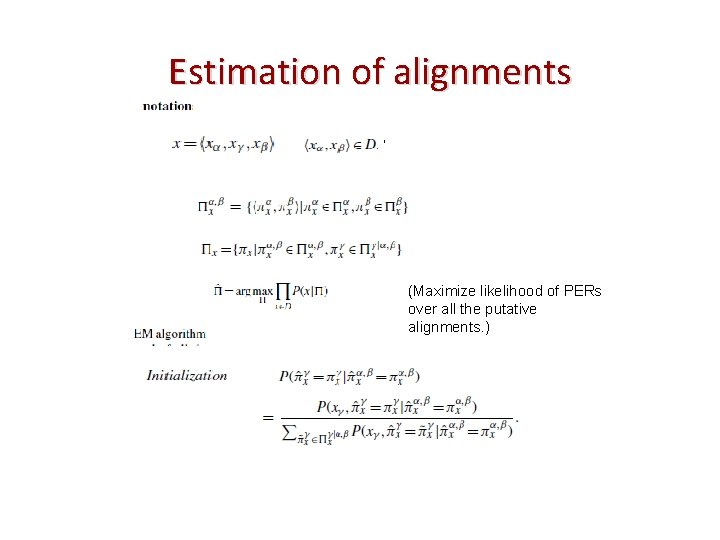
Estimation of alignments , (Maximize likelihood of PERs over all the putative alignments. )

EM continued. . .
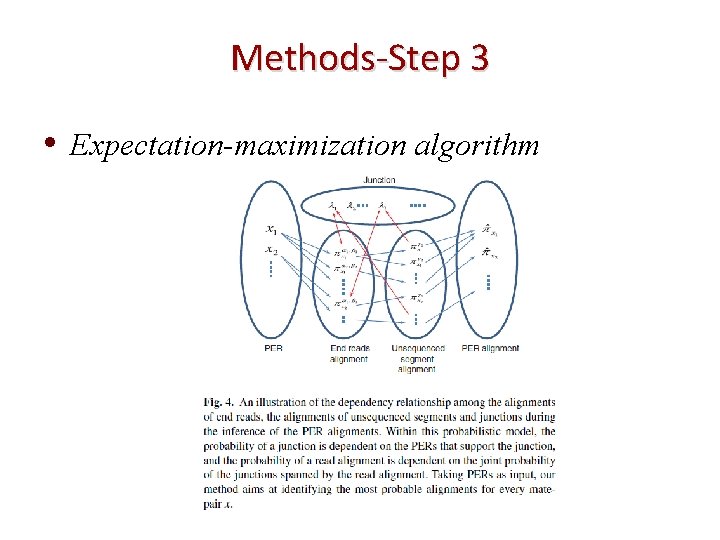
Methods-Step 3 • Expectation-maximization algorithm
Discussion • • Proposed a probabilistic framework to predict the alignment of each PER fragment to a reference genome. By maximizing the likelihood of all PER alignments through a splice graph model Advantageous-higher coverage and specificity than just the alignment of PERs. Capable of detecting trans-chromosome and trans-strand gene fusion events.
Advantages • First, the fragment alignments significantly increase coverage of the transcriptome. Reason: The PER contains almost double information of single read. • Second, it has higher specificity than the junctions in the individual end reads. Reasons: EM algorithm used the information from the entire set of end read alignments.
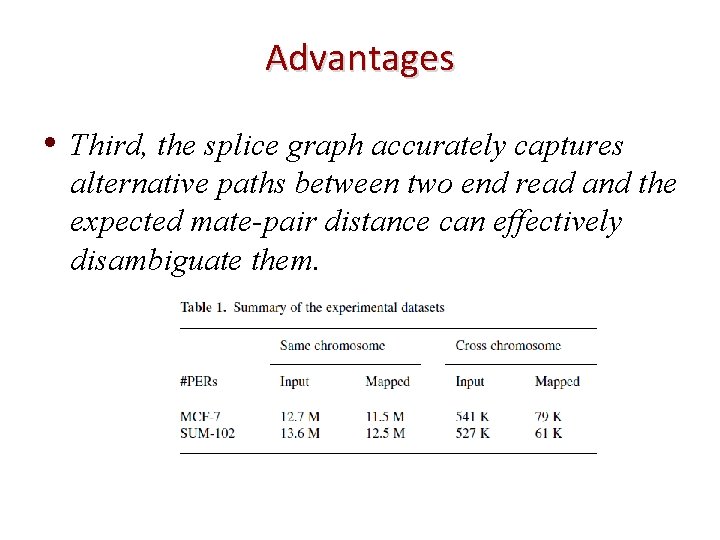
Advantages • Third, the splice graph accurately captures alternative paths between two end read and the expected mate-pair distance can effectively disambiguate them.

Thank you
- Slides: 22