Familial Hypercholesterolemia FH Mechanisms William Cromwell MD Chief







- Slides: 26

Familial Hypercholesterolemia (FH): Mechanisms William Cromwell MD Chief Lipoprotein and Metabolic Disorders Institute Adjunct Associate Professor Wake Forest University School of Medicine FOUNDATION for HEALTH IMPROVEMENT AND TECHNOLOGY

Objectives ► To review ► Classic LDL receptor defects ► Familial Defective Apo. B 100 ► Gain of function PCSK 9 mutations ► Autosomal Recessive FH (Defective LDL Adaptor Protein) ► Deficiency of cholesterol 7 alpha-hydroxylase (CYP 7 A 1) ► Sitosterolemia

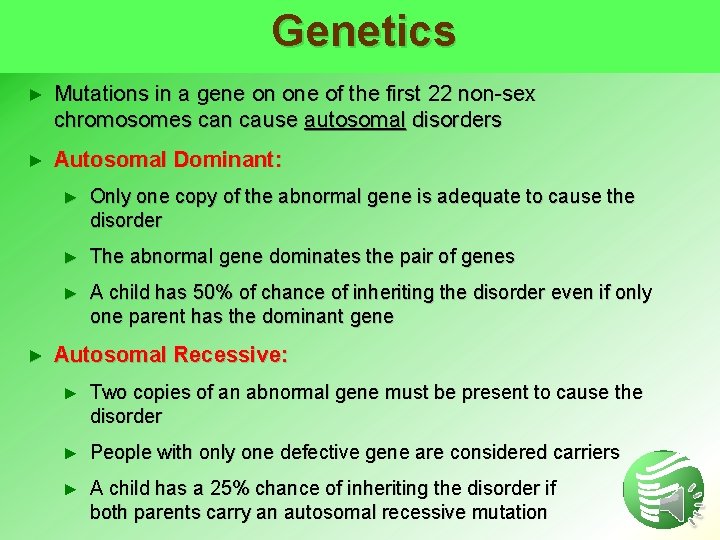
Genetics ► Mutations in a gene on one of the first 22 non-sex chromosomes can cause autosomal disorders ► Autosomal Dominant: ► ► Only one copy of the abnormal gene is adequate to cause the disorder ► The abnormal gene dominates the pair of genes ► A child has 50% of chance of inheriting the disorder even if only one parent has the dominant gene Autosomal Recessive: ► Two copies of an abnormal gene must be present to cause the disorder ► People with only one defective gene are considered carriers ► A child has a 25% chance of inheriting the disorder if both parents carry an autosomal recessive mutation

Familial Hypercholesterolemia (FH) • FH results from defects in the hepatic uptake and degradation of LDL via the LDL-receptor pathway • FH is primarily an autosomal dominant disorder with a genedosage effect. • Dominant etiologies of FH include a loss-of-function mutation in the LDL-receptor gene (LDLR), mutation in the gene encoding apolipoprotein B (APOB), or gain-of-function mutations in PCSK 9. • An autosomal recessive form of FH caused by loss-offunction mutations in LDLRAP 1, which encodes a protein required for clathrin-mediated internalization of the LDL receptor by liver cells, has also been documented.

Monogenic Cholesterol Diseases Disease Prevalence Typical LDL-C (mg/d. L) Familial Hypercholesterolemia 1 per 500* Heterozygous Homozygous Familial Ligand Defective apo. B-100 Heterozygous Homozygous 1 per million 1 per 1000* 270 < 1 per million 320 Autosomal Recessive < 1 per 10 Hypercholesterolemia million** Sitosterolemia * Autosomal Dominant ** Autosomal Recessive 300 650 < 1 per 10 million** 470 100 -600 depending on diet Mutant Gene Mechanism for LDL receptor ↓Function LDL Receptor Nonfunctional receptor Apo. B-100 Decreased binding of LDL to receptors ARH protein Abnormal chaperon protein ABCG 5 and/or ABCG 8* Suppression of receptor gene transcription Goldstein JL & Brown MS. Science 2001: 292: 1310 -1312

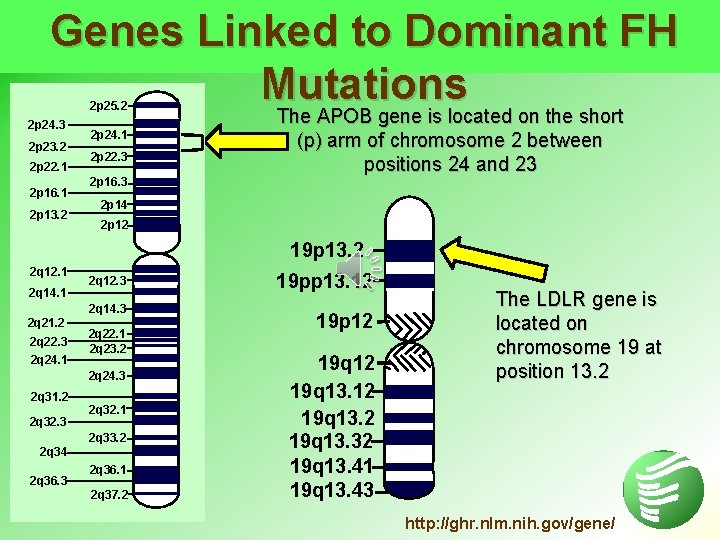
Genes Linked to Dominant FH Mutations The APOB gene is located on the short 2 p 25. 2 2 p 24. 3 2 p 23. 2 2 p 22. 1 2 p 16. 1 2 p 13. 2 2 p 24. 1 2 p 22. 3 2 p 16. 3 The APOB gene is located on the short (p) arm of chromosome 2 between positions 24 and 23 2 p 14 2 p 12 19 p 13. 2 2 q 12. 1 2 q 14. 1 2 q 12. 3 2 q 14. 3 2 q 21. 2 2 q 22. 3 2 q 24. 1 2 q 22. 1 2 q 23. 2 2 q 24. 3 2 q 31. 2 2 q 32. 3 2 q 32. 1 2 q 33. 2 2 q 34 2 q 36. 3 2 q 36. 1 2 q 37. 2 19 pp 13. 12 19 p 12 19 q 13. 32 19 q 13. 41 19 q 13. 43 The LDLR gene is located on chromosome 19 at position 13. 2 http: //ghr. nlm. nih. gov/gene/

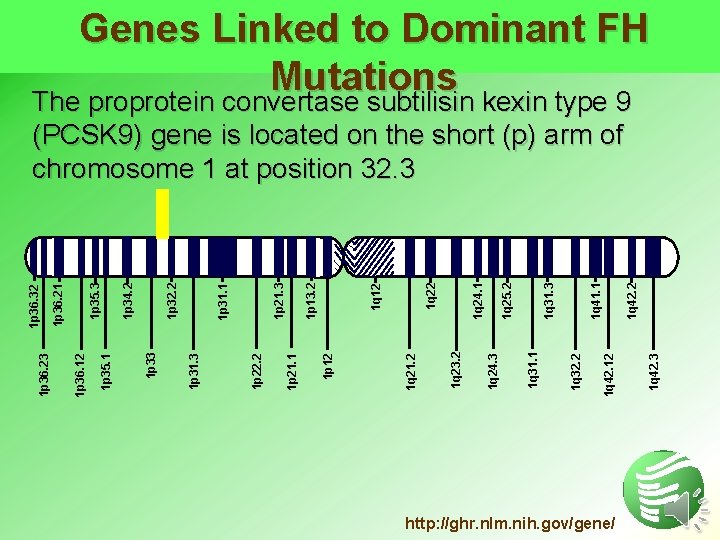
Genes Linked to Dominant FH Mutations The proprotein convertase subtilisin kexin type 9 http: //ghr. nlm. nih. gov/gene/ 1 q 42. 3 1 q 42. 12 1 q 42. 2 1 q 41. 1 1 q 32. 2 1 q 31. 3 1 q 31. 1 1 q 24. 3 1 q 25. 2 1 q 24. 1 1 q 23. 2 1 q 21. 2 1 q 12 1 p 13. 2 1 p 12 1 p 21. 1 1 p 21. 3 1 p 22. 2 1 p 31. 3 1 p 31. 1 1 p 32. 2 1 p 33 1 p 34. 2 1 p 35. 3 1 p 35. 1 1 p 36. 21 1 p 36. 12 1 p 36. 23 1 p 36. 32 (PCSK 9) gene is located on the short (p) arm of chromosome 1 at position 32. 3

The Nobel Prize in Physiology or Medicine 1985 was awarded jointly to Michael S. Brown and Joseph L. Goldstein “for their discoveries concerning the regulation of cholesterol metabolism”

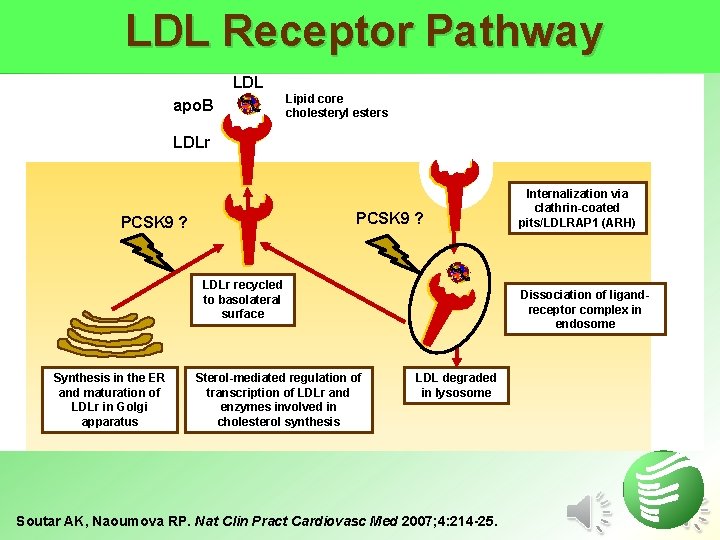
LDL Receptor Pathway LDL apo. B Lipid core cholesteryl esters LDLr PCSK 9 ? LDLr recycled to basolateral surface Synthesis in the ER and maturation of LDLr in Golgi apparatus Sterol-mediated regulation of transcription of LDLr and enzymes involved in cholesterol synthesis Internalization via clathrin-coated pits/LDLRAP 1 (ARH) Dissociation of ligandreceptor complex in endosome LDL degraded in lysosome Soutar AK, Naoumova RP. Nat Clin Pract Cardiovasc Med 2007; 4: 214 -25.

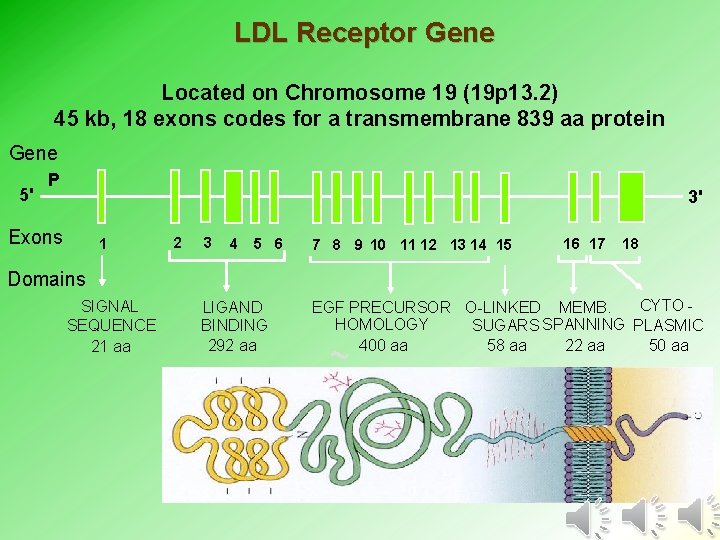
LDL Receptor Gene Located on Chromosome 19 (19 p 13. 2) 45 kb, 18 exons codes for a transmembrane 839 aa protein Gene 5' P 3' Exons 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 Domains SIGNAL SEQUENCE 21 aa LIGAND BINDING 292 aa CYTO EGF PRECURSOR O-LINKED MEMB. HOMOLOGY SUGARS SPANNING PLASMIC 58 aa 50 aa 400 aa 22 aa

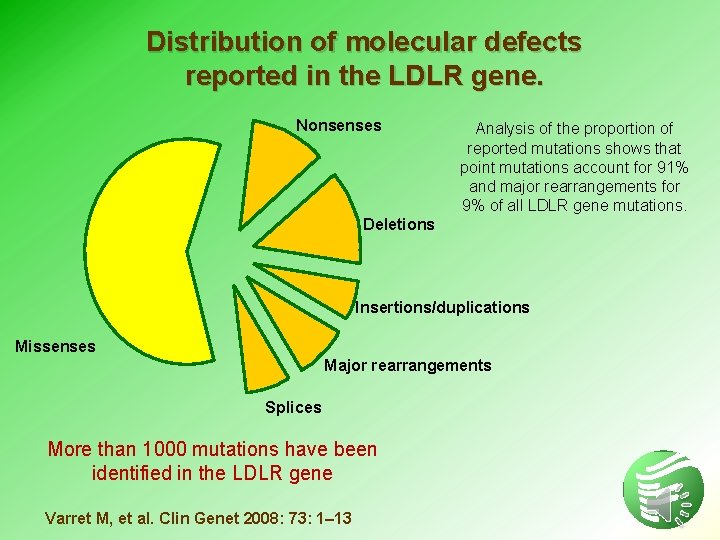
Distribution of molecular defects reported in the LDLR gene. Nonsenses Analysis of the proportion of reported mutations shows that point mutations account for 91% and major rearrangements for 9% of all LDLR gene mutations. Deletions Insertions/duplications Missenses Major rearrangements Splices More than 1000 mutations have been identified in the LDLR gene Varret M, et al. Clin Genet 2008: 73: 1– 13

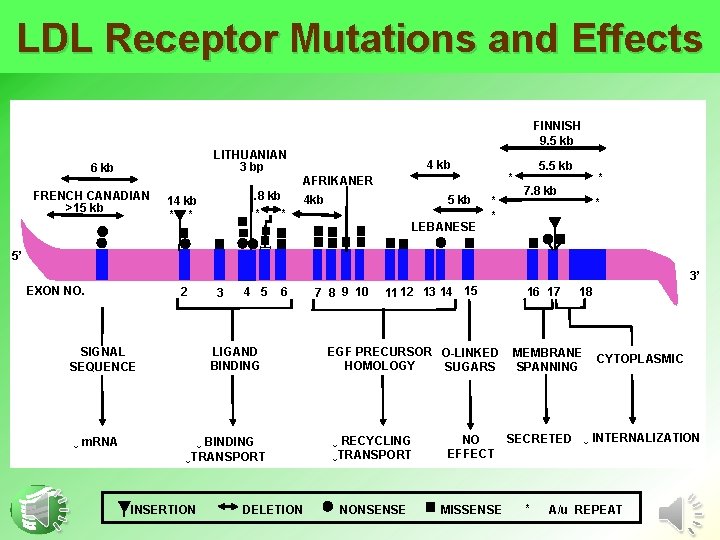
LDL Receptor Mutations and Effects FINNISH 9. 5 kb LITHUANIAN 3 bp 6 kb 4 kb * AFRIKANER FRENCH CANADIAN >15 kb . 8 kb * * 14 kb * * 5 kb 4 kb LEBANESE * * 5. 5 kb * 7. 8 kb * 5’ 3’ EXON NO. 2 4 5 LIGAND BINDING SIGNAL SEQUENCE ↓ m. RNA 3 6 ↓ BINDING ↓TRANSPORT INSERTION DELETION 7 8 9 10 11 12 13 14 15 EGF PRECURSOR O-LINKED HOMOLOGY SUGARS ↓ RECYCLING ↓TRANSPORT NO EFFECT NONSENSE MISSENSE 16 17 18 MEMBRANE SPANNING CYTOPLASMIC SECRETED ↓ INTERNALIZATION * A/u REPEAT

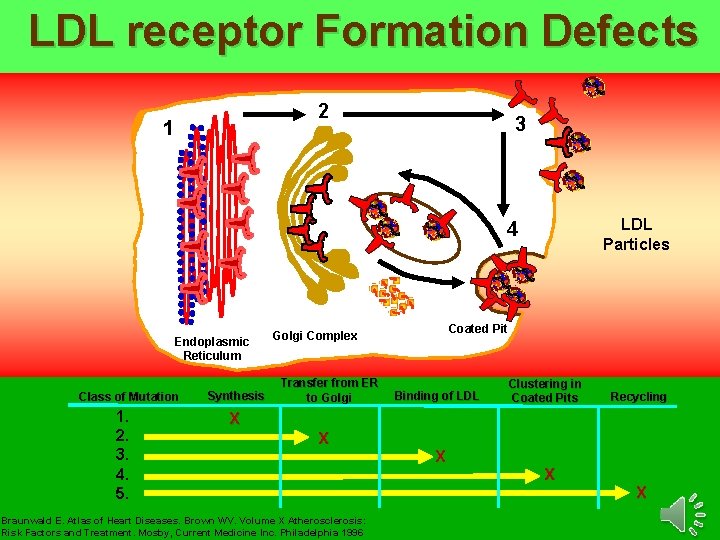
LDL receptor Formation Defects 2 1 3 LDL Particles 4 Endoplasmic Reticulum Class of Mutation 1. 2. 3. 4. 5. Synthesis Coated Pit Golgi Complex Transfer from ER to Golgi Binding of LDL Clustering in Coated Pits Recycling X X Braunwald E. Atlas of Heart Diseases. Brown WV. Volume X Atherosclerosis: Risk Factors and Treatment. Mosby, Current Medicine Inc. Philadelphia 1996 X X X

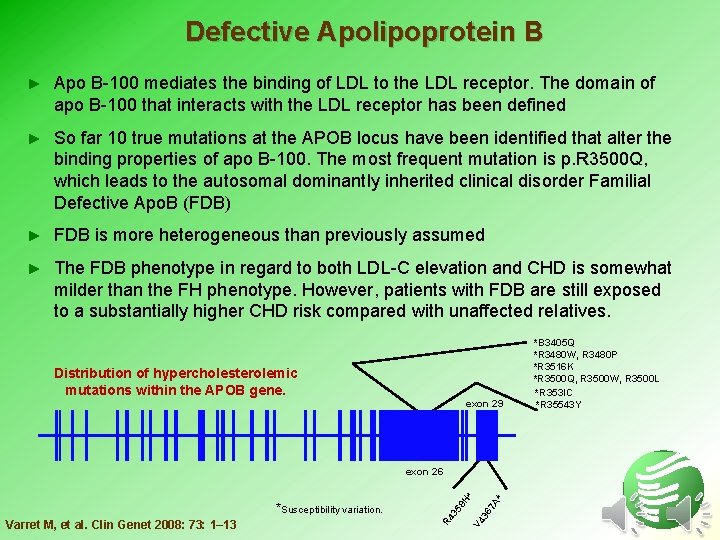
Defective Apolipoprotein B ► Apo B-100 mediates the binding of LDL to the LDL receptor. The domain of apo B-100 that interacts with the LDL receptor has been defined ► So far 10 true mutations at the APOB locus have been identified that alter the binding properties of apo B-100. The most frequent mutation is p. R 3500 Q, which leads to the autosomal dominantly inherited clinical disorder Familial Defective Apo. B (FDB) ► FDB is more heterogeneous than previously assumed ► The FDB phenotype in regard to both LDL-C elevation and CHD is somewhat milder than the FH phenotype. However, patients with FDB are still exposed to a substantially higher CHD risk compared with unaffected relatives. Distribution of hypercholesterolemic mutations within the APOB gene. exon 29 * 7 A 36 V 4 Varret M, et al. Clin Genet 2008: 73: 1– 13 R 4 35 *Susceptibility variation. 8 H * exon 26 *B 3405 Q *R 3480 W, R 3480 P *R 3516 K *R 3500 Q, R 3500 W, R 3500 L *R 353 IC *R 35543 Y

Familial Defective Apo. B Hepatocyte LDL receptors Hepatic Sinusoid ABCA 1 ABCG 4 ABCG 5/8 ABCB 11 Impaired LDL receptor endocytosis of LDL particles carrying defective apo. B-100 Bile Duct

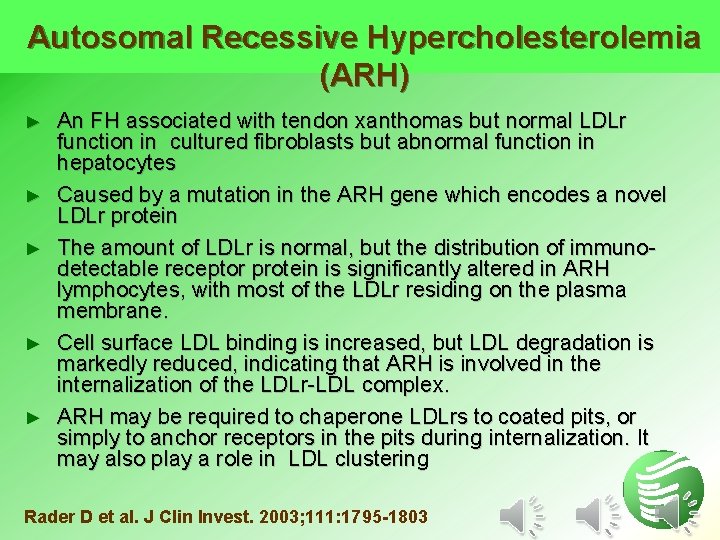
Autosomal Recessive Hypercholesterolemia (ARH) ► ► ► An FH associated with tendon xanthomas but normal LDLr function in cultured fibroblasts but abnormal function in hepatocytes Caused by a mutation in the ARH gene which encodes a novel LDLr protein The amount of LDLr is normal, but the distribution of immunodetectable receptor protein is significantly altered in ARH lymphocytes, with most of the LDLr residing on the plasma membrane. Cell surface LDL binding is increased, but LDL degradation is markedly reduced, indicating that ARH is involved in the internalization of the LDLr-LDL complex. ARH may be required to chaperone LDLrs to coated pits, or simply to anchor receptors in the pits during internalization. It may also play a role in LDL clustering Rader D et al. J Clin Invest. 2003; 111: 1795 -1803

Autosomal Recessive Hypercholesterolemia (ARH) LDL Particle Endocytosis LDL receptor adaptor protein (LDLRAP) functions to bind and help internalize LDL receptors In ARH, LDLRAP is defective, resulting in diminished internalization of LDL particles after binding to LDL receptors. LDL Receptors (LDLr) binding LDL particles Adapted from Rader D et al. J Clin Invest. 2003; 111: 1795 -1803

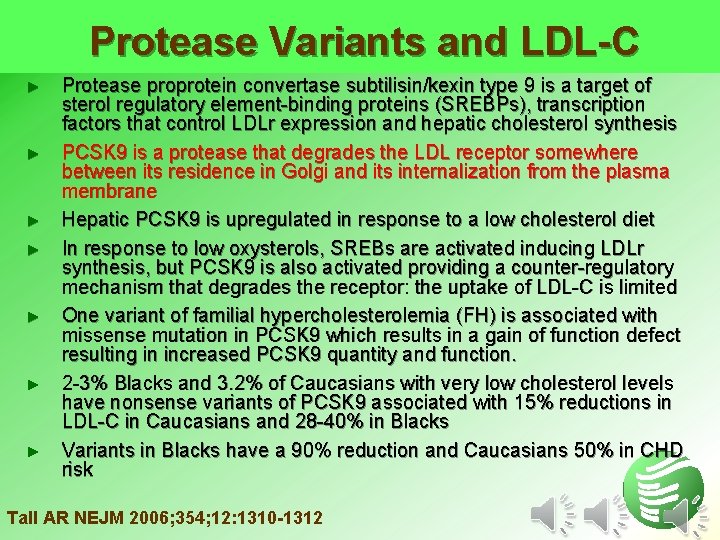
Protease Variants and LDL-C ► ► ► ► Protease proprotein convertase subtilisin/kexin type 9 is a target of sterol regulatory element-binding proteins (SREBPs), transcription factors that control LDLr expression and hepatic cholesterol synthesis PCSK 9 is a protease that degrades the LDL receptor somewhere between its residence in Golgi and its internalization from the plasma membrane Hepatic PCSK 9 is upregulated in response to a low cholesterol diet In response to low oxysterols, SREBs are activated inducing LDLr synthesis, but PCSK 9 is also activated providing a counter-regulatory mechanism that degrades the receptor: the uptake of LDL-C is limited One variant of familial hypercholesterolemia (FH) is associated with missense mutation in PCSK 9 which results in a gain of function defect resulting in increased PCSK 9 quantity and function. 2 -3% Blacks and 3. 2% of Caucasians with very low cholesterol levels have nonsense variants of PCSK 9 associated with 15% reductions in LDL-C in Caucasians and 28 -40% in Blacks Variants in Blacks have a 90% reduction and Caucasians 50% in CHD risk Tall AR NEJM 2006; 354; 12: 1310 -1312

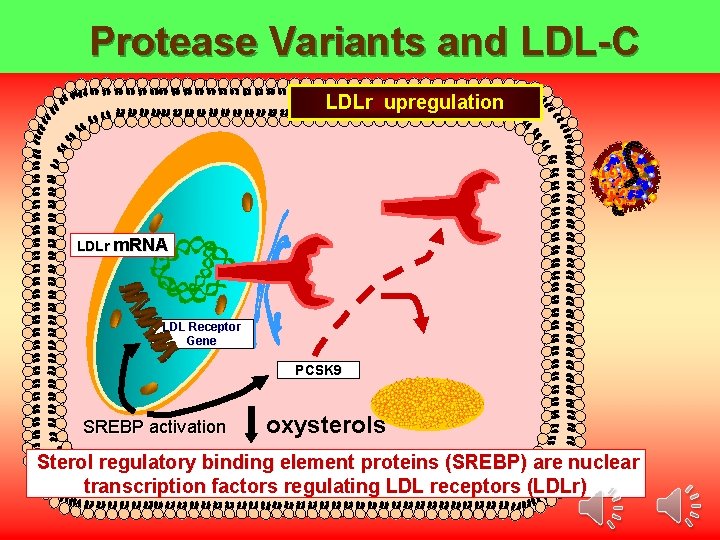
Protease Variants and LDL-C LDLr upregulation LDLr m. RNA LDL Receptor Gene PCSK 9 SREBP activation oxysterols Sterol regulatory binding element proteins (SREBP) are nuclear transcription factors regulating LDL receptors (LDLr) Tall AR NEJM 2006; 354; 12: 1310 -1312

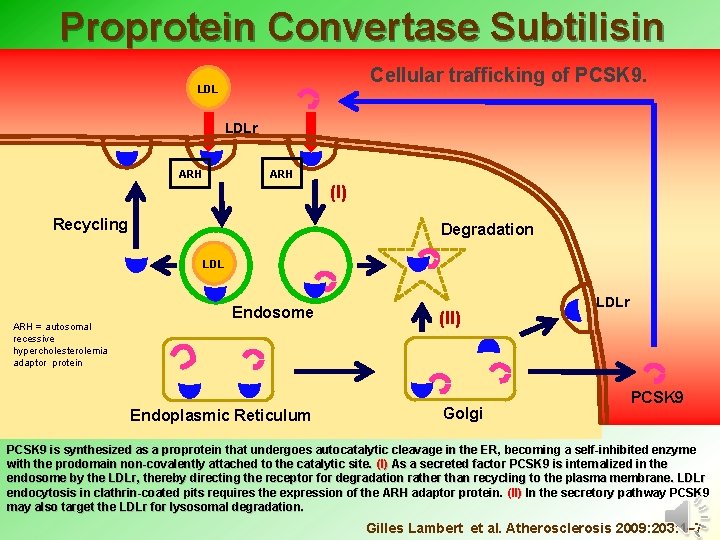
Proprotein Convertase Subtilisin Kexin type 9 Cellular trafficking of PCSK 9. LDLr ARH (I) Recycling Degradation LDL ARH = autosomal recessive hypercholesterolemia adaptor protein Endosome Endoplasmic Reticulum (II) Golgi LDLr PCSK 9 is synthesized as a proprotein that undergoes autocatalytic cleavage in the ER, becoming a self-inhibited enzyme with the prodomain non-covalently attached to the catalytic site. (I) As a secreted factor PCSK 9 is internalized in the endosome by the LDLr, thereby directing the receptor for degradation rather than recycling to the plasma membrane. LDLr endocytosis in clathrin-coated pits requires the expression of the ARH adaptor protein. (II) In the secretory pathway PCSK 9 may also target the LDLr for lysosomal degradation. Gilles Lambert et al. Atherosclerosis 2009: 203: 1– 7

CYP 7 A 1 Cholesterol 7 Alpha Hydroxylase Deficiency ► Described as an autosomal co-dominant disorder affecting cholesterol 7α-hydroxylase activity, the first enzyme in the classical pathway for bile acid synthesis ► A block in the conversion of cholesterol to bile acids by this rate-limiting enzyme blocks the secretion of cholesterol to bile from the liver, increases hepatic cholesterol and a downregulation of the LDL receptor Pullinger, et al J. Clin. Invest. 2002; 110: 109 -117 Kwiterovich PO, ed. The Johns Hopkins textbook of dyslipidemia. 1 st ed. Philadelphia, PA: Lippincott Williams & Wilkins; 2009

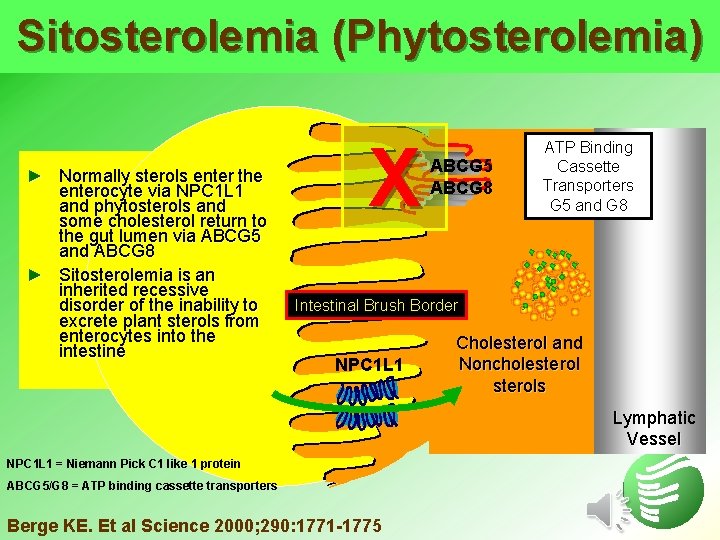
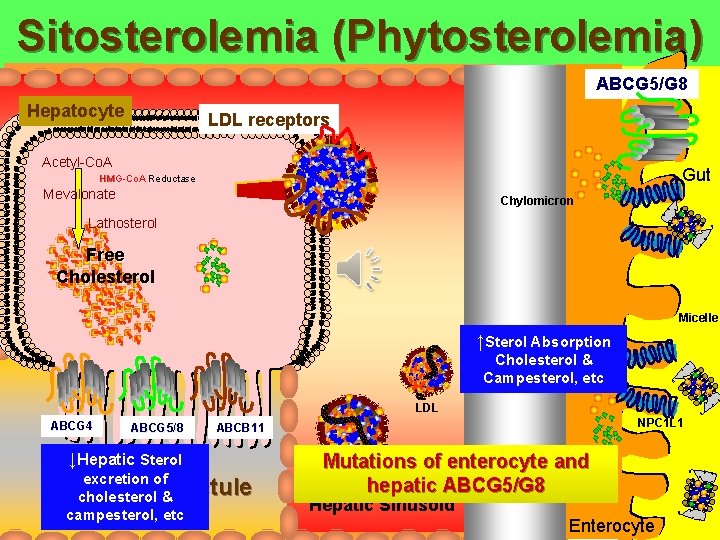
Sitosterolemia (Phytosterolemia) ► Normally sterols enter the enterocyte via NPC 1 L 1 and phytosterols and some cholesterol return to the gut lumen via ABCG 5 and ABCG 8 ► Sitosterolemia is an inherited recessive disorder of the inability to excrete plant sterols from enterocytes into the intestine X ABCG 5 ABCG 8 ATP Binding Cassette Transporters G 5 and G 8 Intestinal Brush Border NPC 1 L 1 Cholesterol and Noncholesterols Lymphatic Vessel NPC 1 L 1 = Niemann Pick C 1 like 1 protein ABCG 5/G 8 = ATP binding cassette transporters Berge KE. Et al Science 2000; 290: 1771 -1775

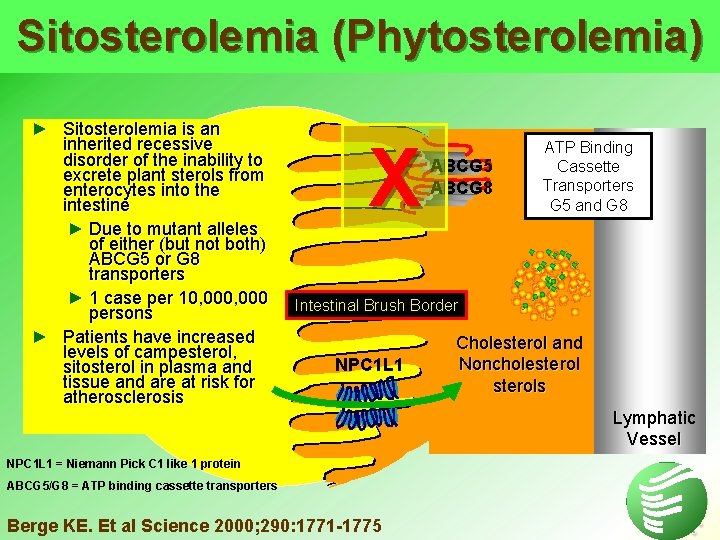
Sitosterolemia (Phytosterolemia) ► Sitosterolemia is an inherited recessive disorder of the inability to excrete plant sterols from enterocytes into the intestine ► Due to mutant alleles of either (but not both) ABCG 5 or G 8 transporters ► 1 case per 10, 000 persons ► Patients have increased levels of campesterol, sitosterol in plasma and tissue and are at risk for atherosclerosis X ABCG 5 ABCG 8 ATP Binding Cassette Transporters G 5 and G 8 Intestinal Brush Border NPC 1 L 1 = Niemann Pick C 1 like 1 protein ABCG 5/G 8 = ATP binding cassette transporters Berge KE. Et al Science 2000; 290: 1771 -1775 Cholesterol and Noncholesterols Lymphatic Vessel

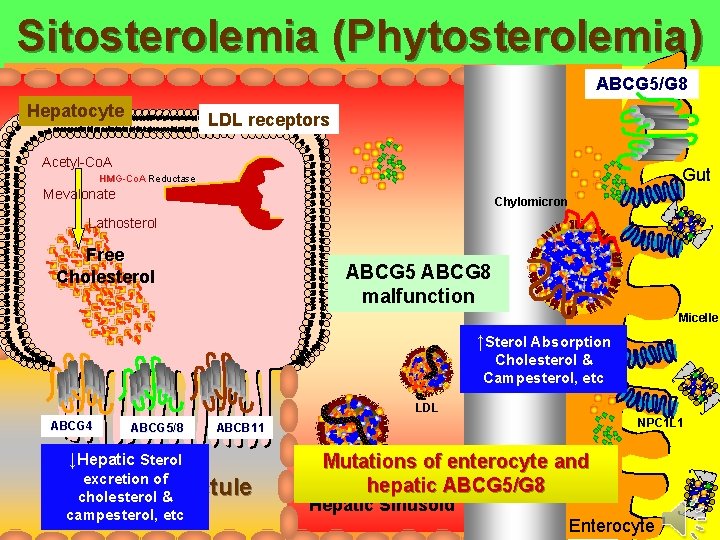
Sitosterolemia (Phytosterolemia) ABCG 5/G 8 Hepatocyte LDL receptors Acetyl-Co. A Gut HMG-Co. A Reductase Mevalonate Chylomicron Lathosterol Free Cholesterol ABCG 5 ABCG 8 malfunction Micelle ↑Sterol Absorption Cholesterol & Campesterol, etc LDL ABCG 4 ABCG 5/8 ↓Hepatic Sterol excretion of cholesterol & campesterol, etc NPC 1 L 1 ABCB 11 Bile Ductule Mutations VLDL of enterocyte and Lacteal hepatic ABCG 5/G 8 Hepatic Sinusoid Enterocyte

Sitosterolemia (Phytosterolemia) ABCG 5/G 8 Hepatocyte LDL receptors Acetyl-Co. A Gut HMG-Co. A Reductase Mevalonate Chylomicron Lathosterol Free Cholesterol Micelle ↑Sterol Absorption Cholesterol & Campesterol, etc LDL ABCG 4 ABCG 5/8 ↓Hepatic Sterol excretion of cholesterol & campesterol, etc NPC 1 L 1 ABCB 11 Bile Ductule Mutations VLDL of enterocyte and Lacteal hepatic ABCG 5/G 8 Hepatic Sinusoid Enterocyte

Thank you for listening FOUNDATION for HEALTH IMPROVEMENT AND TECHNOLOGY
 Sitosterolemia
Sitosterolemia Lesson 3 commander in chief and chief diplomat
Lesson 3 commander in chief and chief diplomat Gestion de patrimoine familial
Gestion de patrimoine familial Familial hypocalciuric hypercalcemia
Familial hypocalciuric hypercalcemia Edward syndrome
Edward syndrome Familial roots
Familial roots Dr pattan casper wy
Dr pattan casper wy Familial hypertriglyceridemia
Familial hypertriglyceridemia Monoploidy
Monoploidy Danette format familial
Danette format familial Robertsonian translocation
Robertsonian translocation Familial hypocalciuric hypercalcemia
Familial hypocalciuric hypercalcemia Cromwell
Cromwell Was oliver cromwell a hero or villian
Was oliver cromwell a hero or villian Oliver cromwell hero
Oliver cromwell hero Oliver cromwell
Oliver cromwell Robert moray
Robert moray Wdl bioinformatics
Wdl bioinformatics Oliver cromwell
Oliver cromwell Was oliver cromwell a hero or villian
Was oliver cromwell a hero or villian Cromwell hero or villain
Cromwell hero or villain What did stephanie cromwell and elizabeth venko decide?
What did stephanie cromwell and elizabeth venko decide? Curentul romantic
Curentul romantic Protectoratul lui oliver cromwell
Protectoratul lui oliver cromwell Salisbury eskü
Salisbury eskü Oliver cromwell
Oliver cromwell Mechanisms of evolution
Mechanisms of evolution