Factors Affecting Enzyme Activity Enzymes are large globular

Factors Affecting Enzyme Activity
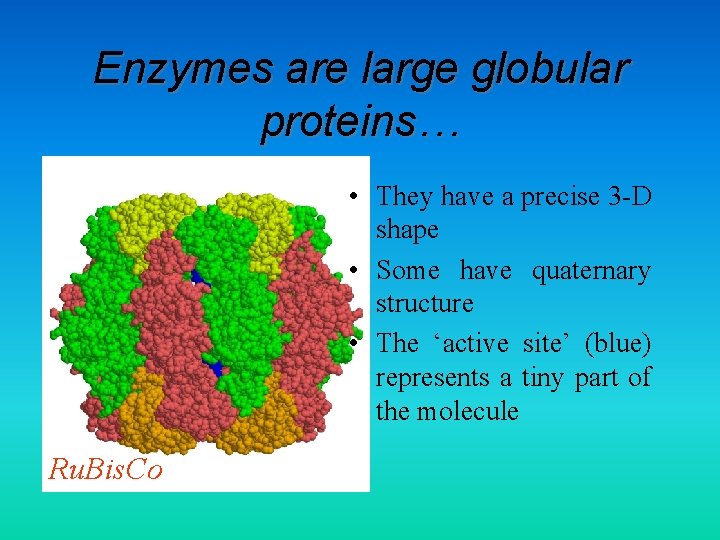
Enzymes are large globular proteins… • They have a precise 3 -D shape • Some have quaternary structure • The ‘active site’ (blue) represents a tiny part of the molecule Ru. Bis. Co
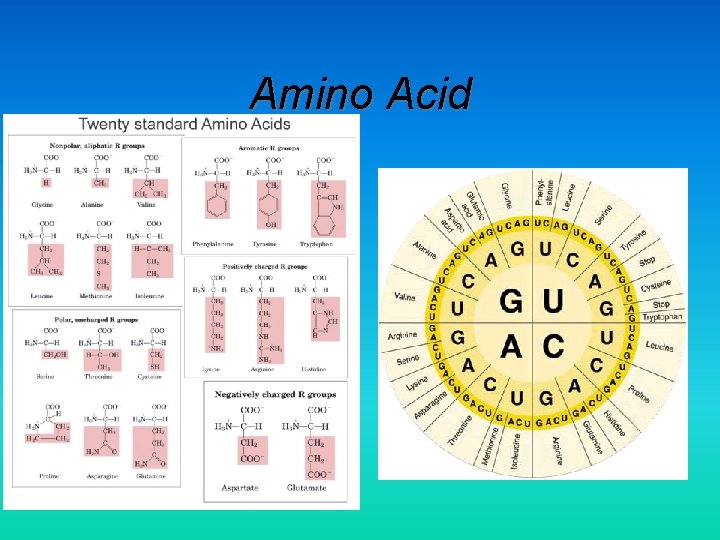
Amino Acid
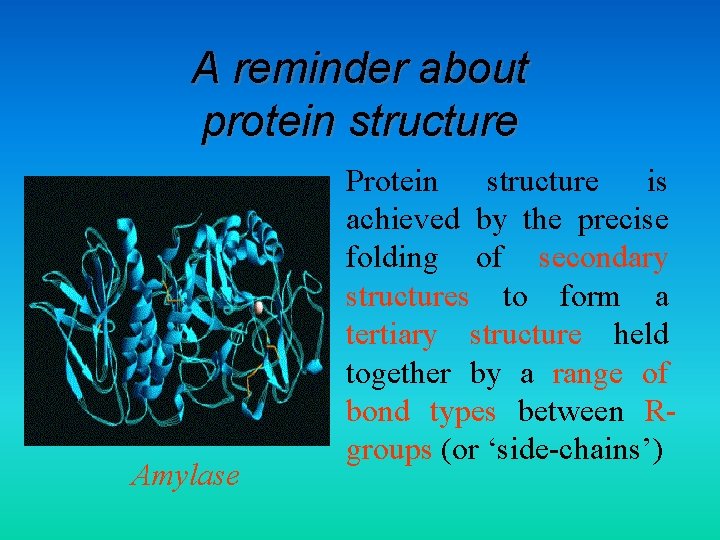
A reminder about protein structure Amylase • Protein structure is achieved by the precise folding of secondary structures to form a tertiary structure held together by a range of bond types between Rgroups (or ‘side-chains’)
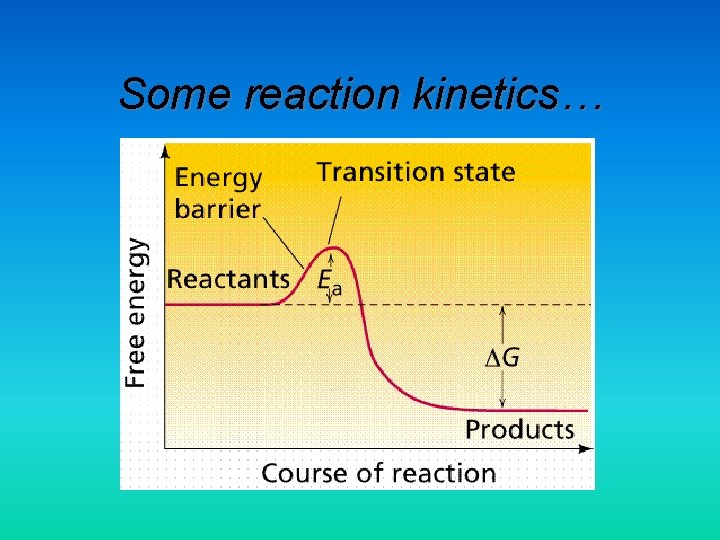
Some reaction kinetics…
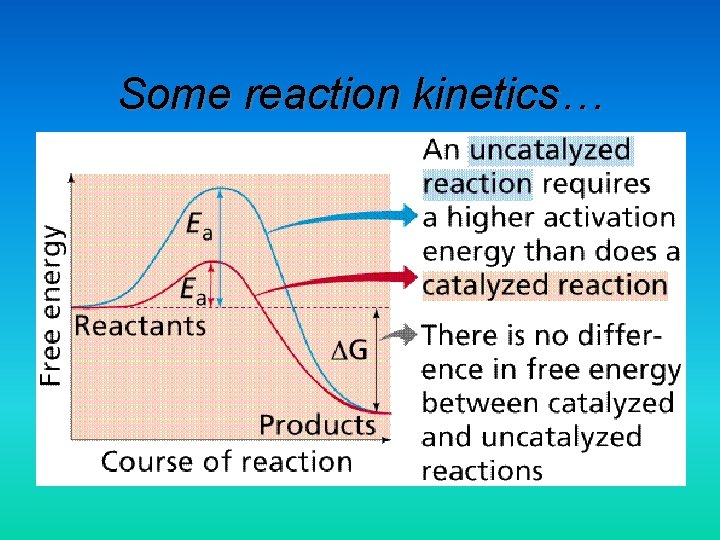
Some reaction kinetics…
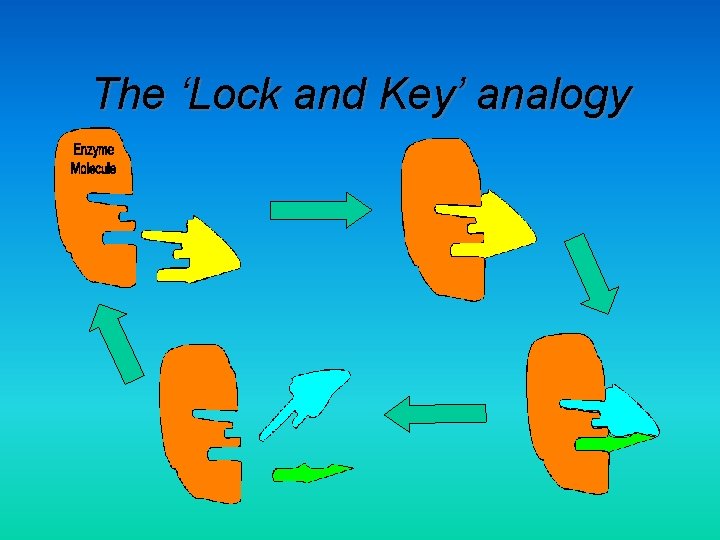
The ‘Lock and Key’ analogy
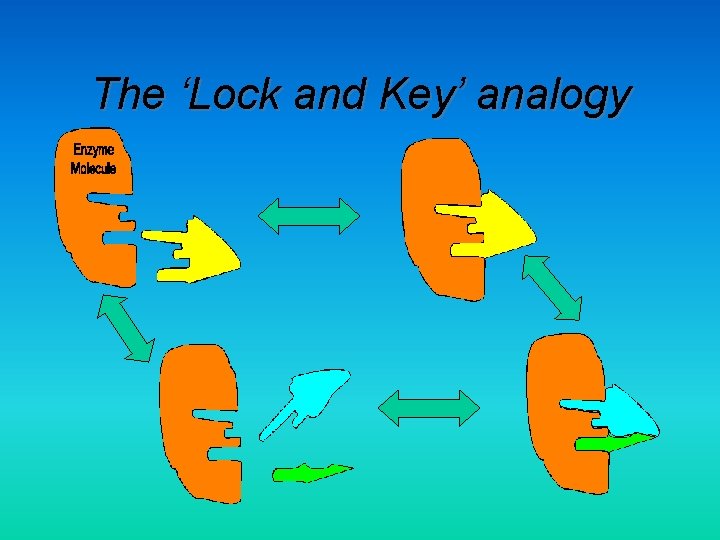
The ‘Lock and Key’ analogy
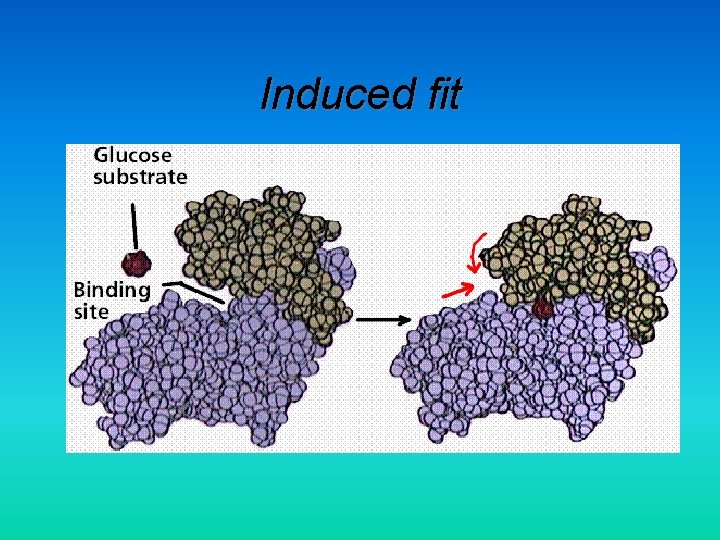
Induced fit
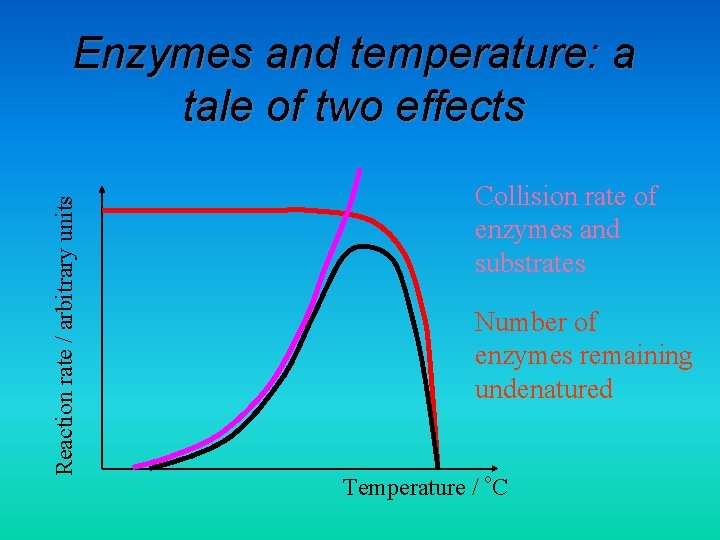
Reaction rate / arbitrary units Enzymes and temperature: a tale of two effects Collision rate of enzymes and substrates Number of enzymes remaining undenatured Temperature / o. C
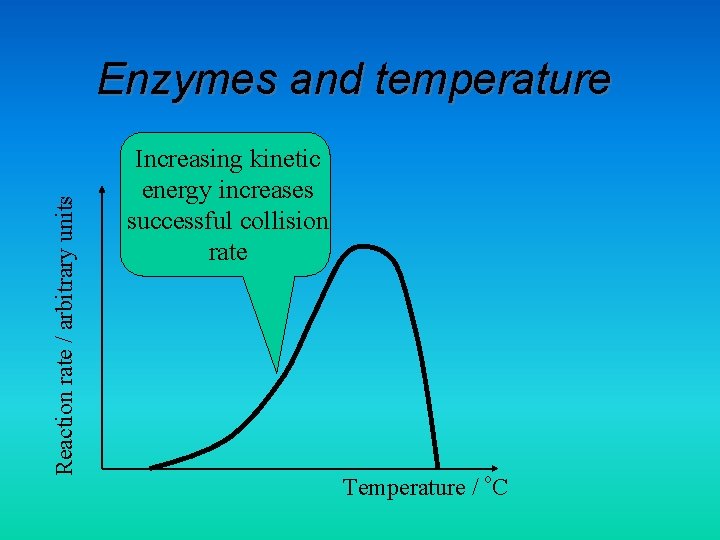
Reaction rate / arbitrary units Enzymes and temperature Increasing kinetic energy increases successful collision rate Temperature / o. C
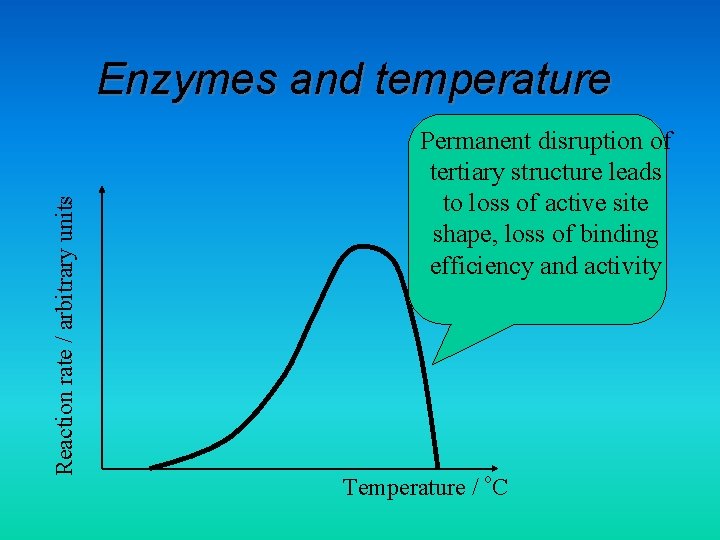
Reaction rate / arbitrary units Enzymes and temperature Permanent disruption of tertiary structure leads to loss of active site shape, loss of binding efficiency and activity Temperature / o. C
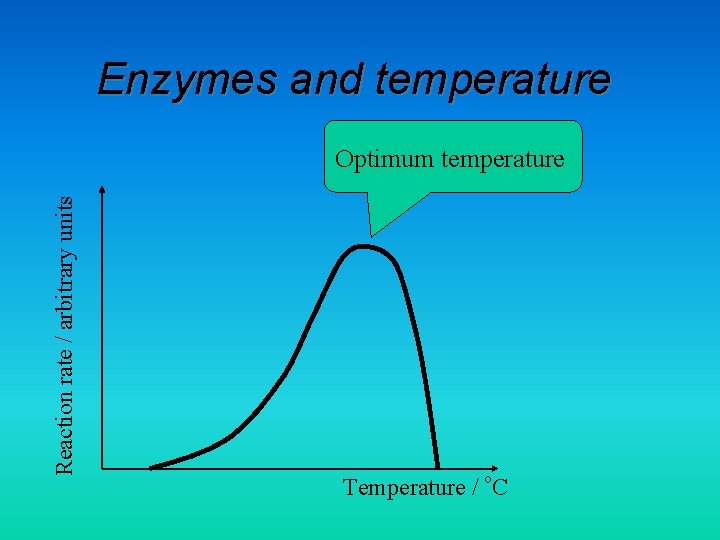
Enzymes and temperature Reaction rate / arbitrary units Optimum temperature Temperature / o. C
Enzymes and p. H • The precise shape of an enzyme (and hence its active site) depends on the tertiary structure of the protein • Tertiary structure is held together by weak bonds (including hydrogen bonds) between R-groups (or ‘side-chains’) • Changing p. H can cause these side chains to ionise resulting in the loss of H-bonding…
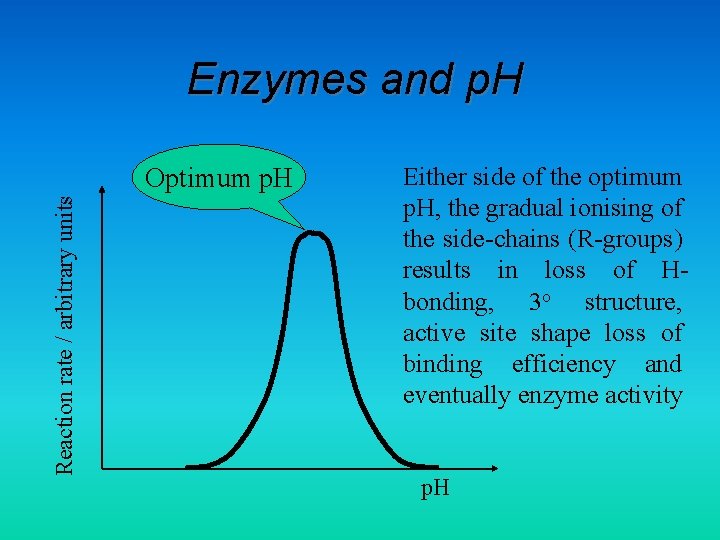
Enzymes and p. H Reaction rate / arbitrary units Optimum p. H Either side of the optimum p. H, the gradual ionising of the side-chains (R-groups) results in loss of Hbonding, 3 o structure, active site shape loss of binding efficiency and eventually enzyme activity p. H
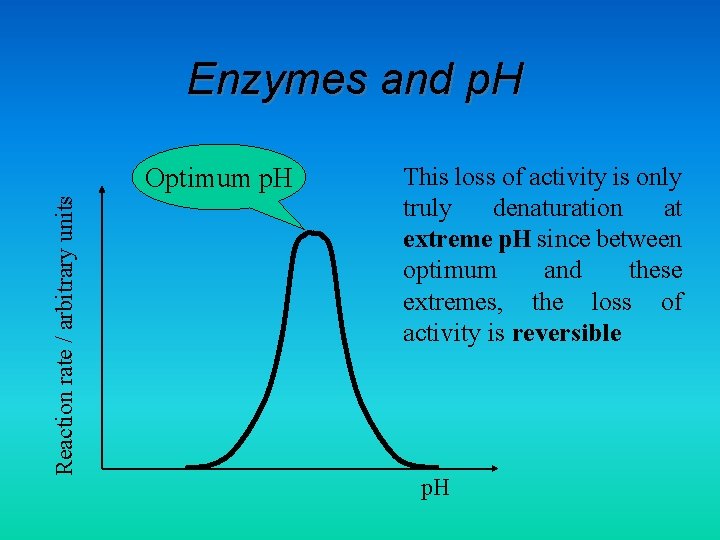
Enzymes and p. H Reaction rate / arbitrary units Optimum p. H This loss of activity is only truly denaturation at extreme p. H since between optimum and these extremes, the loss of activity is reversible p. H
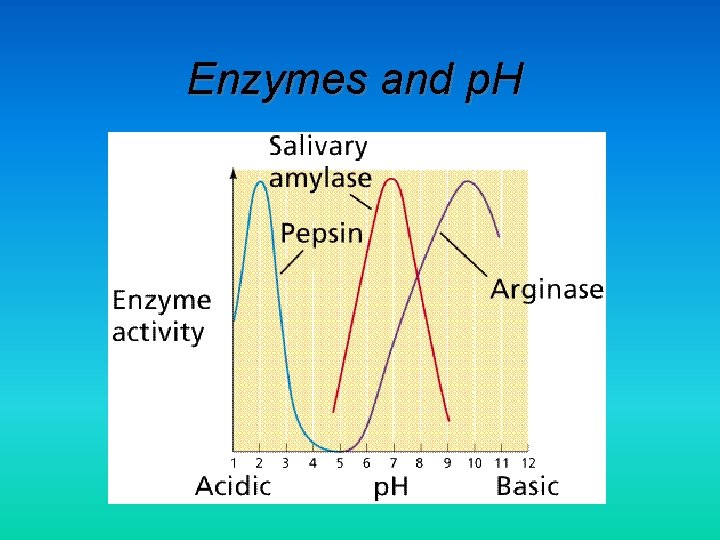
Enzymes and p. H
![Initial reaction rate / arbitrary units Enzymes and [S] As soon as a reaction Initial reaction rate / arbitrary units Enzymes and [S] As soon as a reaction](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-18.jpg)
Initial reaction rate / arbitrary units Enzymes and [S] As soon as a reaction begins, [S] begins to fall and so it is important that initial reaction rates are measured [S]
![Initial reaction rate / arbitrary units Enzymes and [S] Initial reaction rate / arbitrary units Enzymes and [S]](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-19.jpg)
Initial reaction rate / arbitrary units Enzymes and [S]
![Initial reaction rate / arbitrary units Enzymes and [S] Increasing [S] increases collision rate Initial reaction rate / arbitrary units Enzymes and [S] Increasing [S] increases collision rate](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-20.jpg)
Initial reaction rate / arbitrary units Enzymes and [S] Increasing [S] increases collision rate and increases reaction rate [S]
![Initial reaction rate / arbitrary units Enzymes and [S] All active sites are occupied. Initial reaction rate / arbitrary units Enzymes and [S] All active sites are occupied.](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-21.jpg)
Initial reaction rate / arbitrary units Enzymes and [S] All active sites are occupied. Enzymes are working at maximum rate. All active sites are not occupied [S]
![Initial reaction rate / arbitrary units Enzymes and [S] Maximum turnover number or Vmax Initial reaction rate / arbitrary units Enzymes and [S] Maximum turnover number or Vmax](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-22.jpg)
Initial reaction rate / arbitrary units Enzymes and [S] Maximum turnover number or Vmax has been reached [S]
![Initial reaction rate / arbitrary units Enzymes and [enzyme] Can we explain this in Initial reaction rate / arbitrary units Enzymes and [enzyme] Can we explain this in](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-23.jpg)
Initial reaction rate / arbitrary units Enzymes and [enzyme] Can we explain this in terms of the proportions of active sites occupied? What factor is limiting here? [Enzyme]

Enzymes and inhibitors • Inhibitors are molecules that prevent enzymes reaching their maximum turnover numbers • Some inhibitors compete with the substrate Active site directed inhibition for the active site • Some inhibitors affect the active site shape Non-active siteenzyme directed inhibition by binding to the elsewhere on the enzyme
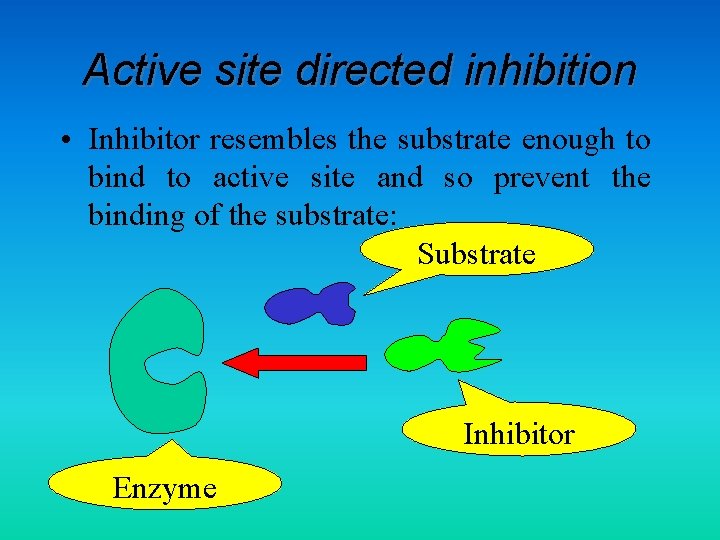
Active site directed inhibition • Inhibitor resembles the substrate enough to bind to active site and so prevent the binding of the substrate: Substrate Inhibitor Enzyme
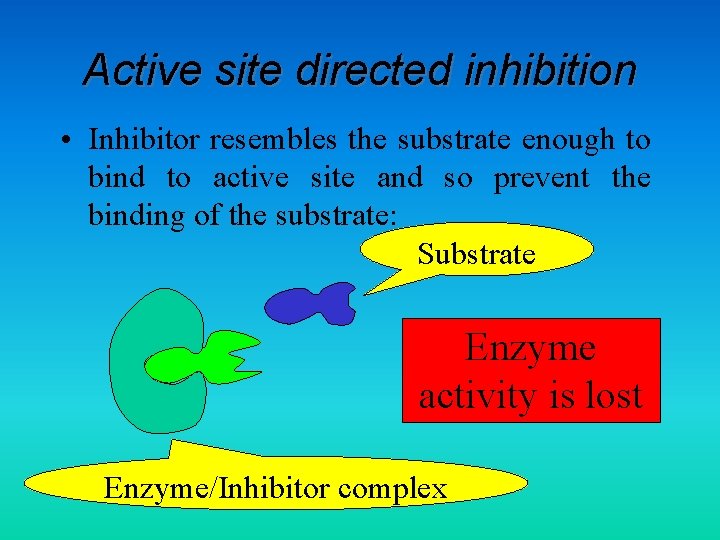
Active site directed inhibition • Inhibitor resembles the substrate enough to bind to active site and so prevent the binding of the substrate: Substrate Enzyme activity is lost Enzyme/Inhibitor complex
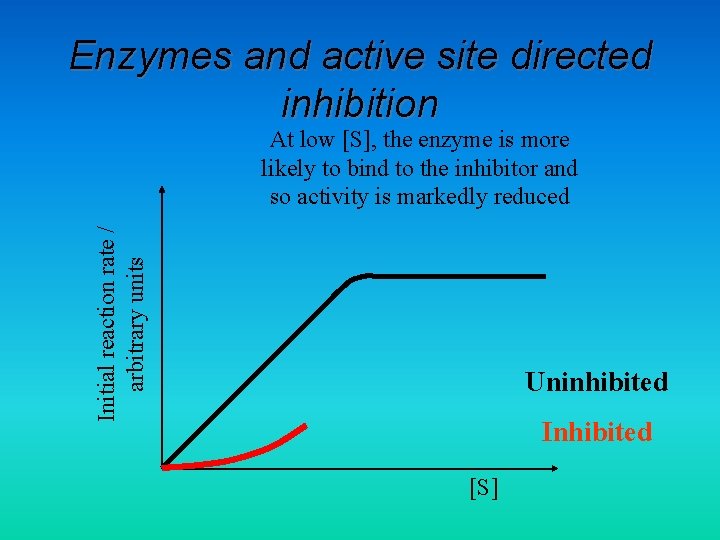
Enzymes and active site directed inhibition Initial reaction rate / arbitrary units At low [S], the enzyme is more likely to bind to the inhibitor and so activity is markedly reduced Uninhibited Inhibited [S]
![Enzymes and active site directed inhibition Initial reaction rate / arbitrary units As [S] Enzymes and active site directed inhibition Initial reaction rate / arbitrary units As [S]](http://slidetodoc.com/presentation_image_h/a3d3b737c2db18d165271253dd9b2d5d/image-28.jpg)
Enzymes and active site directed inhibition Initial reaction rate / arbitrary units As [S] rises, the enzyme is increasingly likely to bind to the substrate and so activity increases Uninhibited Inhibited [S]
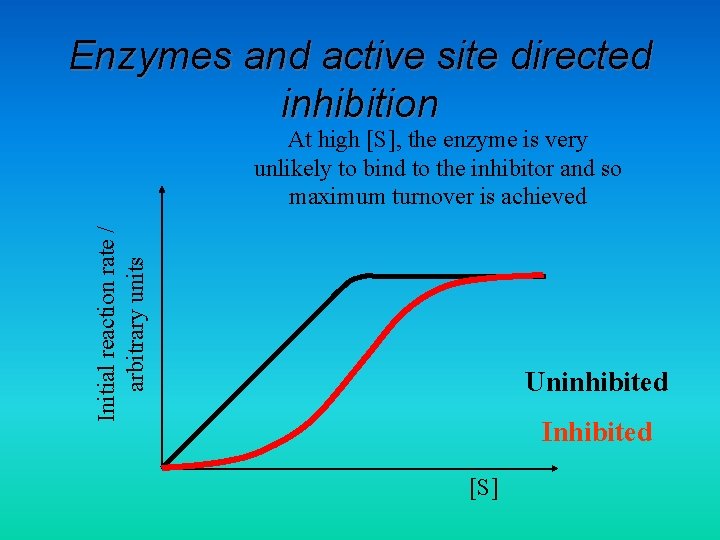
Enzymes and active site directed inhibition Initial reaction rate / arbitrary units At high [S], the enzyme is very unlikely to bind to the inhibitor and so maximum turnover is achieved Uninhibited Inhibited [S]
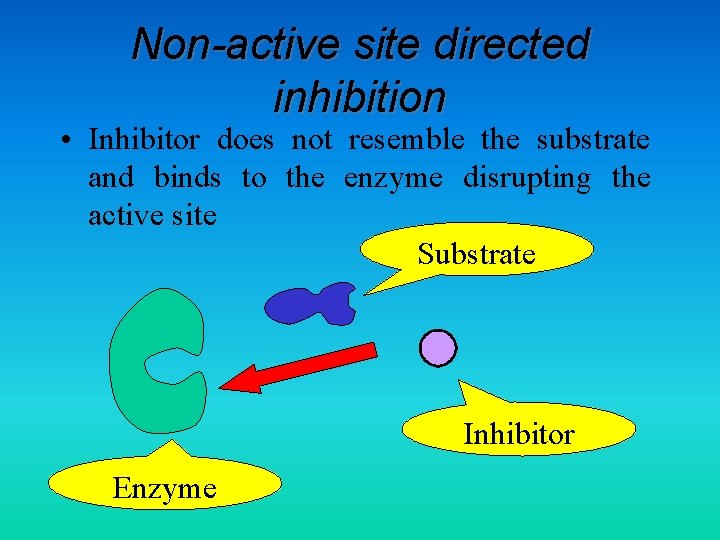
Non-active site directed inhibition • Inhibitor does not resemble the substrate and binds to the enzyme disrupting the active site Substrate Inhibitor Enzyme
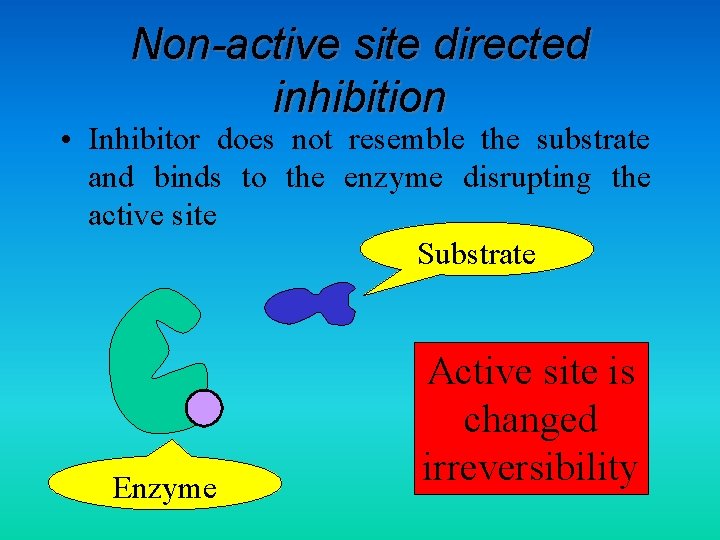
Non-active site directed inhibition • Inhibitor does not resemble the substrate and binds to the enzyme disrupting the active site Substrate Enzyme Active site is changed irreversibility
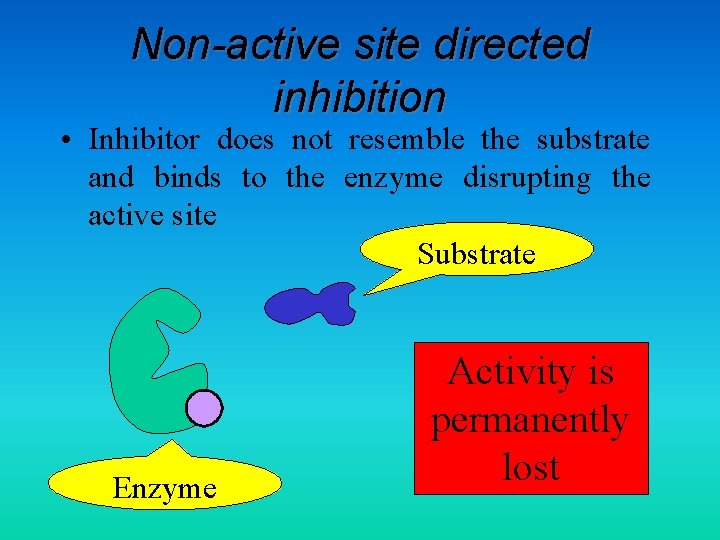
Non-active site directed inhibition • Inhibitor does not resemble the substrate and binds to the enzyme disrupting the active site Substrate Enzyme Activity is permanently lost
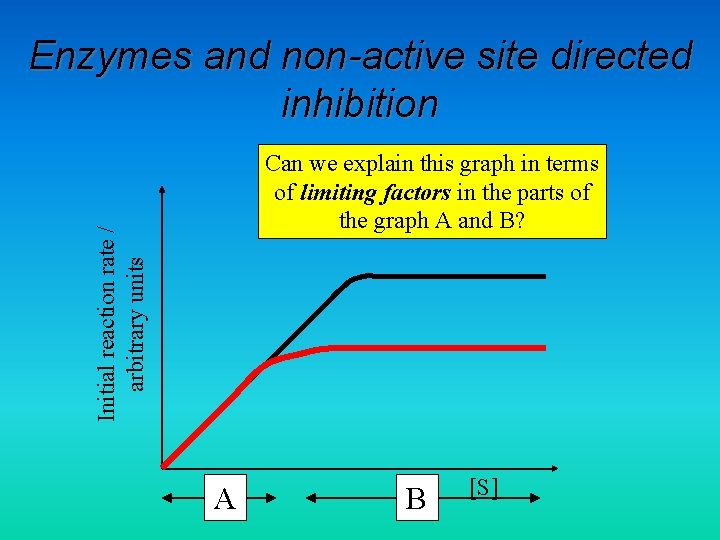
Enzymes and non-active site directed inhibition Initial reaction rate / arbitrary units Can we explain this graph in terms of limiting factors in the parts of the graph A and B? A B [S]
- Slides: 33