Facilitate Scientific Data Sharing by Sharing Informatics Tools

Facilitate Scientific Data Sharing by Sharing Informatics Tools and Standards Second Meeting of the Board on Research Data and Information September 24, 2009 Belinda Seto and James Luo National Institute of Biomedical Imaging and Bioengineering National Institutes of Health

NIH Data Sharing Policy NIH believes that data sharing is essential for expedited translation of research results into knowledge, products, and procedures to improve human health. The policy reaffirmed the principle that data should be made as widely and freely available as possible while safeguarding the privacy of research participants, and protecting confidential and proprietary data.
NIH Bioinformatics Initiatives l NIH GWAS - Genome Wide Association Study l ca. BIG The Cancer Biomedical Grid The goal- of these initiatives is to. Informatics build infrastructure and networks to facilitate data sharing, integration, l BIRN - The Biomedical Informatics Research Network and interoperability. l CTSA - Clinical and Translational Science Awards l NIH Blueprint Neuroimaging Informatics Softwares are open source and free to download. l NCBC - National Centers for Biomedical Computing
NIH Bioinformatics Initiatives l NIH GWAS - Genome Wide Association Study - db. Ga. P l ca. BIG - The Cancer Biomedical Informatics Grid l BIRN - The Biomedical Informatics Research Network l CTSA - Clinical and Translational Science Awards l NIH Blueprint Neuroimaging Informatics - NITRC l NCBC - National Centers for Biomedical Computing - i 2 b 2 l The above trans-NIH infrastructures, tools and standards were presented at 3 rd US-China Roundtable on Scientific Data Cooperation. - NBIA, Rembrandt

l Impact and benefit of sharing tools l– 2 case studies

NIH Blueprint – NITRC l NITRC - Neuroimaging Informatics Tools and Resources Clearinghouse: A web site and a community l NITRC helps research laboratories to share their NIHfunded neuroimaging tools and resources. – To provide the neuroimaging informatics tools and resources to the neuroimaging research community at large – To provide opportunities for public comment regarding neuroimaging informatics tools and resources by the neuroimaging research community at large l NITRC identifies software, data sets and other resources developed under NIH grants useful to the greater community and encourages their developers to share them.

NITRC Results l Within 1. 5 years since its first release, NITRC has – hosted 220 tools and resources – more than 53% of the tools on NITRC are new tools that have not been previously shared online. – built a community of 6, 000 unique visitors per month – 1, 077+ registered users (11% non-English) – with 42, 000 downloads l With an average tool development grant of $350, 000 it is estimated that if 6% of the tools on NITRC today are utilized by another research laboratory instead of that laboratory requesting new government funding, this project will have more than paid for itself.
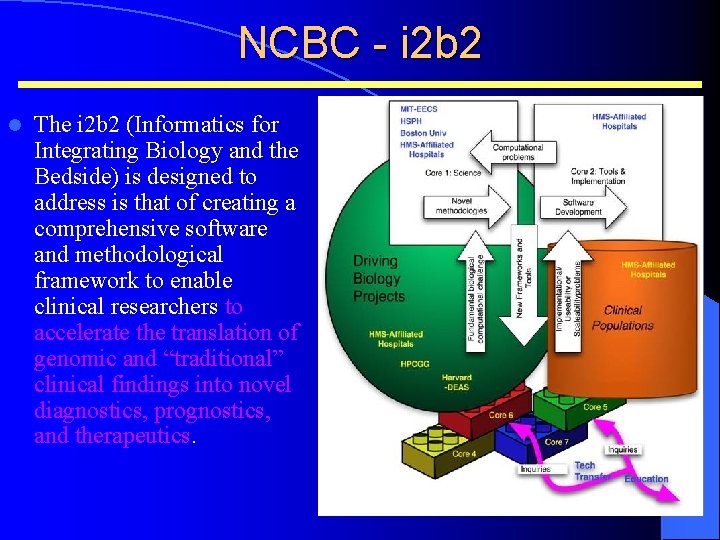
NCBC - i 2 b 2 l The i 2 b 2 (Informatics for Integrating Biology and the Bedside) is designed to address is that of creating a comprehensive software and methodological framework to enable clinical researchers to accelerate the translation of genomic and “traditional” clinical findings into novel diagnostics, prognostics, and therapeutics.
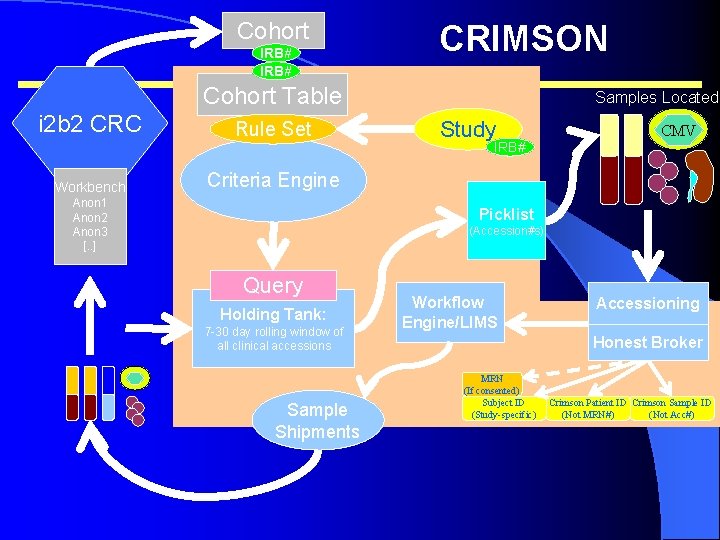
Cohort IRB# CRIMSON Cohort Table i 2 b 2 CRC Workbench Rule Set Samples Located Study CMV IRB# Criteria Engine Anon 1 Anon 2 Anon 3 [. . ] Picklist (Accession#s) Query Holding Tank: 7 -30 day rolling window of all clinical accessions Sample Shipments Workflow Engine/LIMS Accessioning Honest Broker MRN (If consented) Subject ID (Study-specific) Crimson Patient ID Crimson Sample ID (Not MRN#) (Not Acc#)
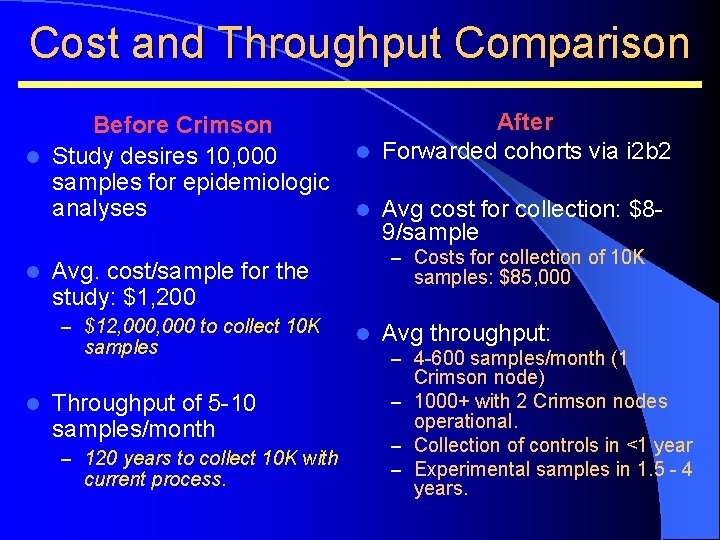
Cost and Throughput Comparison Before Crimson l Study desires 10, 000 samples for epidemiologic analyses l l samples Throughput of 5 -10 samples/month – 120 years to collect 10 K with current process. Avg cost for collection: $89/sample – Costs for collection of 10 K Avg. cost/sample for the study: $1, 200 – $12, 000 to collect 10 K l After l Forwarded cohorts via i 2 b 2 samples: $85, 000 l Avg throughput: – 4 -600 samples/month (1 Crimson node) – 1000+ with 2 Crimson nodes operational. – Collection of controls in <1 year – Experimental samples in 1. 5 - 4 years.

Looking Forward l Outcomes of 3 rd US-China Roundtable meeting – Dr. Huixiong John Zhang, University of Electronic Science and Technology of China (UESTC): l Interest in leveraging NIH bioinformatics infrastructure and initiatives, e. g. ca. BIG, BIRN, CTSA, NCBC (i 2 b 2), etc. to facilitate data sharing – Dr. Xuan Dong, First Hospital of Chiang Zhou City: l Identified two MRI imaging data sets and time series neuro-physiological data sets for consideration for sharing. l NBIA will be used as the tools to share the image data. l Physio. Net will be used as the tools to share the neuro-physiological data

Looking Forward l Met with Drs. Yixue Li and Lei Liu, Shanghai Center for Bioinformation Technology and discussed potential collaborations on data and standards sharing: – Clinical research informatics and sharing of standards (including HL 7, IHE, DICOM, etc. ) – Medical imaging, data sharing and decision support. – GWAS informatics and database, data analysis, data standards.

Driving toward tangible outcomes l Develop demonstration projects from China and U. S. toward scientific data sharing l Share data standards l Share experience with electronic medical records
- Slides: 13