F sarahfriMV Multivariate Linkage and Association Sarah Medland
























- Slides: 51

F: sarahfri_MV

Multivariate Linkage and Association Sarah Medland Manuel Ferreira -with heavy borrowing from Kate Morley and Frühling Rijsdijk

MV analyses can address ¢ Questions of common aetiology Same gene (snp) l Co-incidental covariation due to LD between two different genes l Co-variation due to shared social/environmental risk factors l Aust. Albatross

MV analyses can address ¢ Questions of common aetiology Same gene (snp) l Co-incidental covariation due to LD between two different genes l Co-variation due to shared social/environmental risk factors l ¢ Pleiotropy occurs when a single gene influences multiple phenotypic traits. Bilby

Studying multiple phenotypes… ¢ Run multiple univariate analyses on correlated traits Bandicoot


Studying multiple phenotypes… ¢ Run multiple univariate analyses l Correct for multiple testing… • Bonferroni • Correction for equivalent number of independent variables l Doesn’t really address the idea of common aetiology Blue ringed octopus

Studying multiple phenotypes… ¢ Run multiple univariate analyses l Try and determine if the coincident linkage/association is statistically unlikely Box jellyfish

Studying multiple phenotypes… ¢ Run multiple univariate analyses l Try and determine if the coincident linkage/association is statistically unlikely • Simulate/Permute data and assess how often this group of traits reaches this pattern of sig. by chance Brown Snake

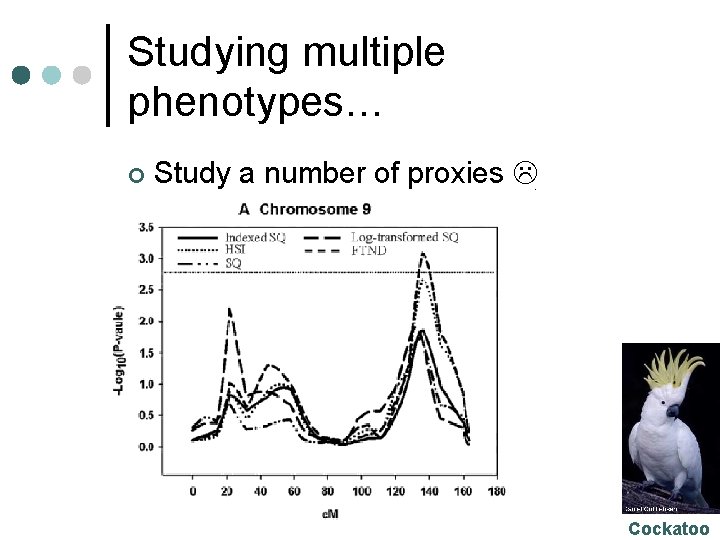
Studying multiple phenotypes… ¢ Study a number of proxies Cockatoo

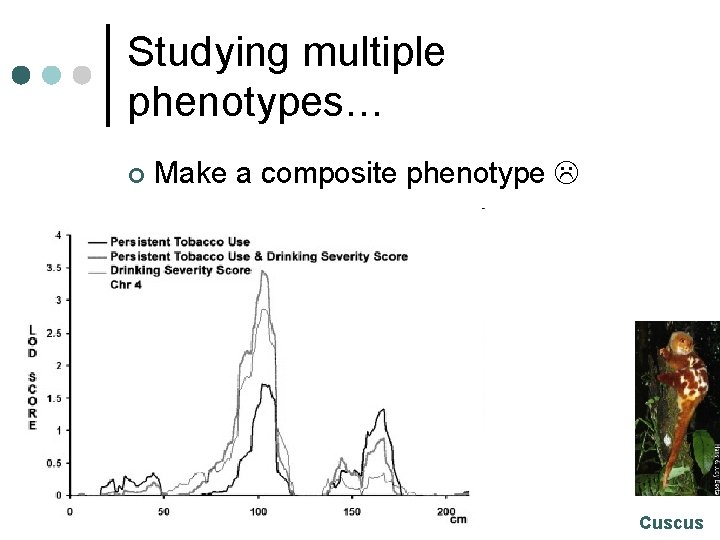
Studying multiple phenotypes… ¢ Make a composite phenotype Cuscus

Studying multiple phenotypes… ¢ Make a factor score combine both factor level and traitspecific effects l latent factor effects are inherently pleiotropic l residual effects are not l assumes factor loadings are constant across genome l Cuscus (again)

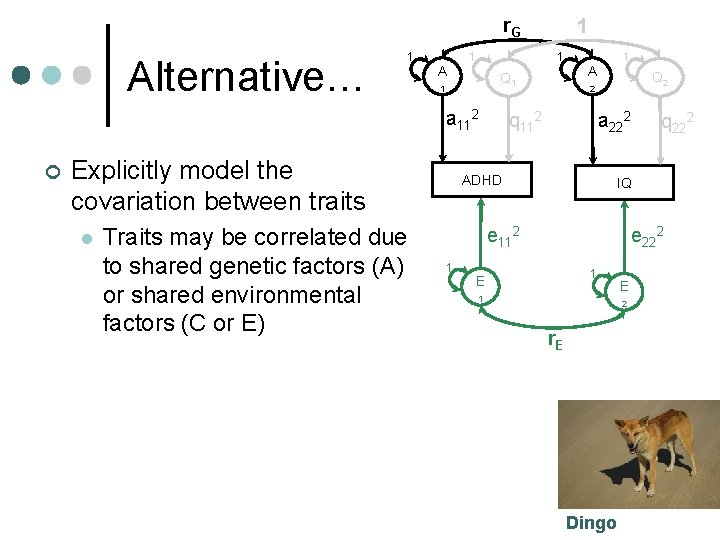
r. G Alternative… 1 A 1 1 Q 1 1 a 112 ¢ Explicitly model the covariation between traits l Traits may be correlated due to shared genetic factors (A) or shared environmental factors (C or E) 1 1 A Q 2 2 q 112 a 222 ADHD IQ e 112 1 q 222 e 222 1 E 2 r. E Dingo

Explicitly model the covariation between traits… Directly assess pleiotropy ¢ Increased power ¢ l Esp. when the pattern of QTL effects is different from the background covariation • ie positive r but gene effects are in opposite directions ¢ Reduced multiple testing Drop Bear

Multivariate analysis… ¢ In the context of linkage analysis when we have family data two traits measured in twin pairs l Interested in: l • Cross-trait covariance within individuals • Cross-trait covariance between twins • MZ: DZ ratio of cross-trait covariance between twins Echidna

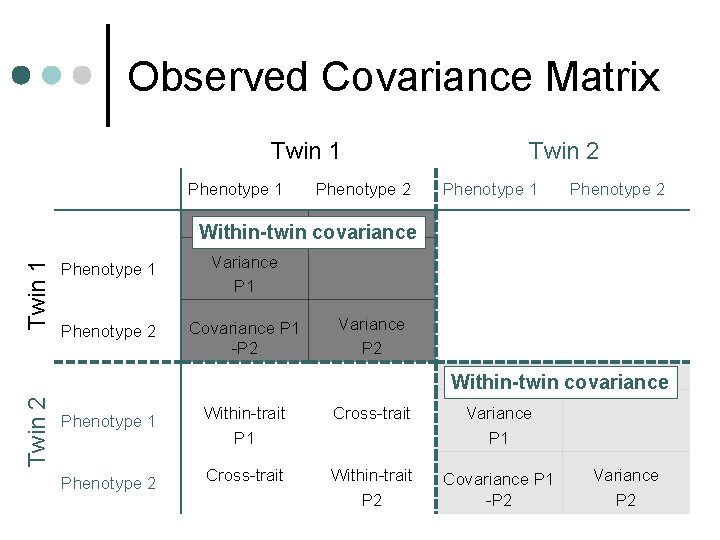
Observed Covariance Matrix Twin 1 Twin 2 Twin 1 Phenotype 2 Twin 2 Phenotype 1 Variance P 1 Phenotype 2 Covariance P 1 -P 2 Variance P 2 Phenotype 1 Within-trait P 1 Cross-trait Variance P 1 Phenotype 2 Cross-trait Within-trait P 2 Covariance P 1 -P 2 Phenotype 2 Variance P 2

Observed Covariance Matrix Twin 1 Phenotype 2 Twin 2 Phenotype 1 Phenotype 2 Twin 1 Within-twin covariance Phenotype 1 Variance P 1 Phenotype 2 Covariance P 1 -P 2 Variance P 2 Twin 2 Within-twin covariance Phenotype 1 Within-trait P 1 Cross-trait Variance P 1 Phenotype 2 Cross-trait Within-trait P 2 Covariance P 1 -P 2 Variance P 2

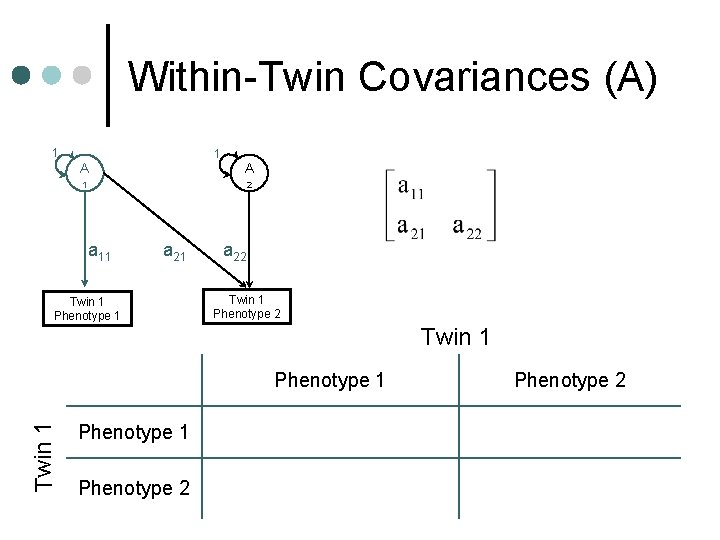
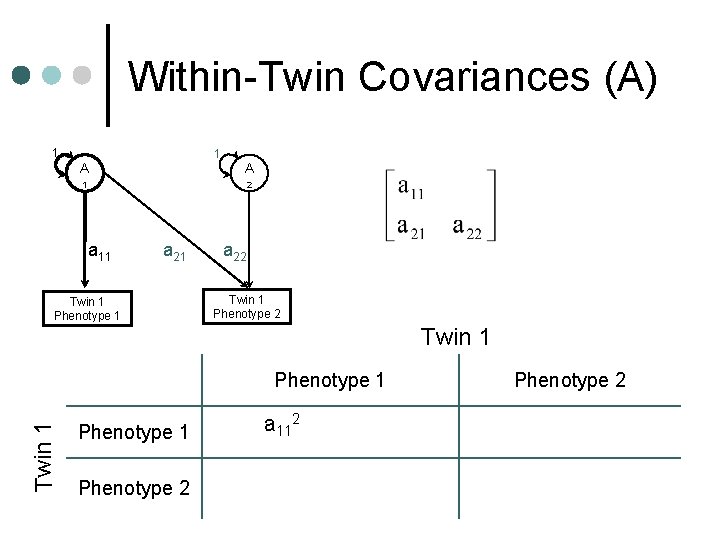
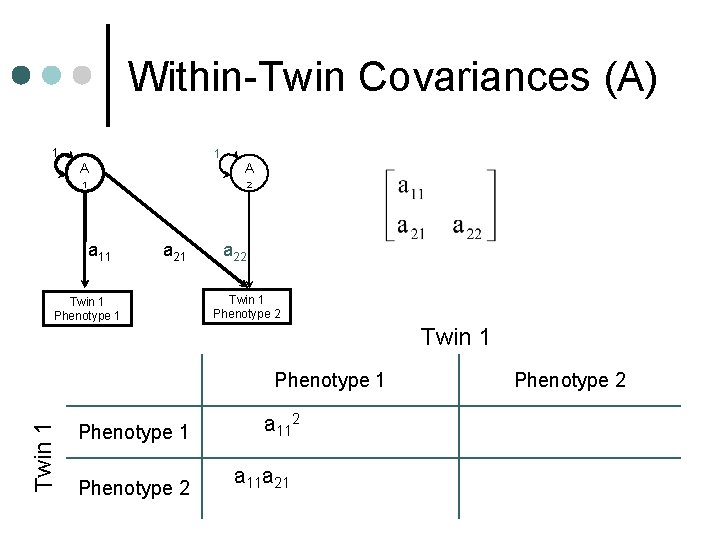
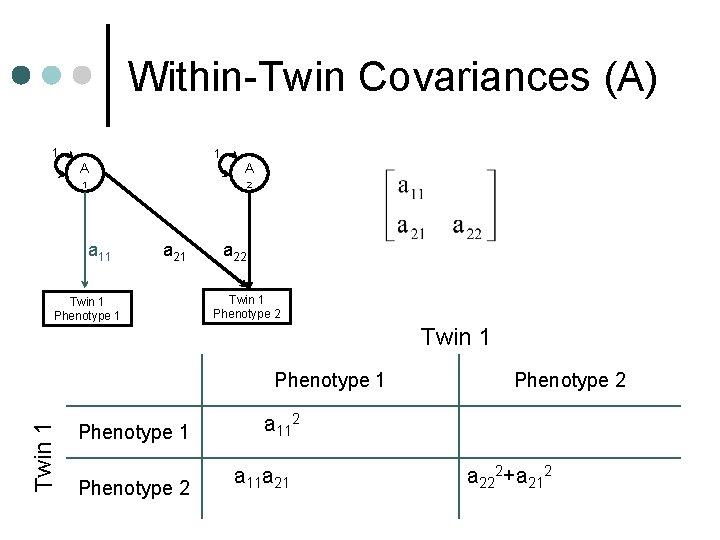
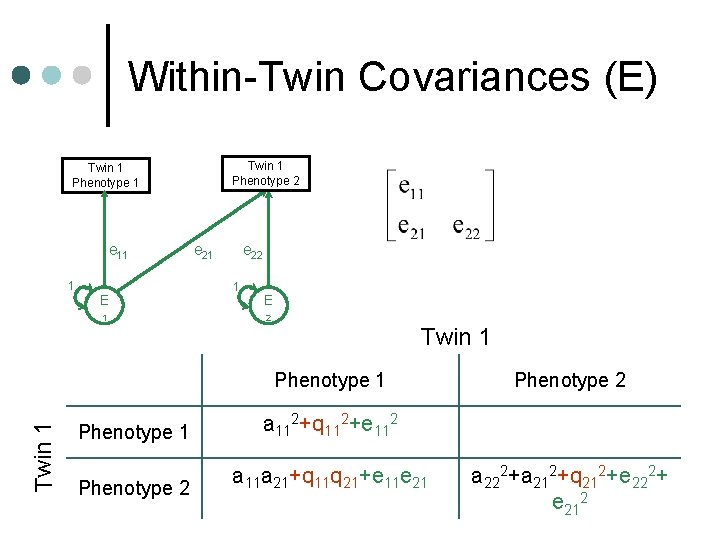
Within-Twin Covariances (A) 1 1 A A 1 2 a 11 a 21 Twin 1 Phenotype 1 a 22 Twin 1 Phenotype 1 a 112+c 112+e 112 Phenotype 2 a 11 a 21+c 11 c 21+e 11 e 21 Phenotype 2 a 222+a 212+c 222+c 212+e 2 2 22 +e 21

Within-Twin Covariances (A) 1 1 A A 1 2 a 11 a 21 Twin 1 Phenotype 1 a 22 Twin 1 Phenotype 1 a 112+c 112+e 112 Phenotype 2 a 11 a 21+c 11 c 21+e 11 e 21 Phenotype 2 a 222+a 212+c 222+c 212+e 2 2 22 +e 21

Within-Twin Covariances (A) 1 1 A A 1 2 a 11 a 21 Twin 1 Phenotype 1 a 22 Twin 1 Phenotype 1 a 112+c 112+e 112 Phenotype 2 a 11 a 21+c 11 c 21+e 11 e 21 Phenotype 2 a 222+a 212+c 222+c 212+e 2 2 22 +e 21

Within-Twin Covariances (A) 1 1 A A 1 2 a 11 a 21 Twin 1 Phenotype 1 a 22 Twin 1 Phenotype 1 a 112+c 112+e 112 Phenotype 2 a 11 a 21+c 11 c 21+e 11 e 21 Phenotype 2 a 222+a 212+c 222+c 212+e 2 2 22 +e 21

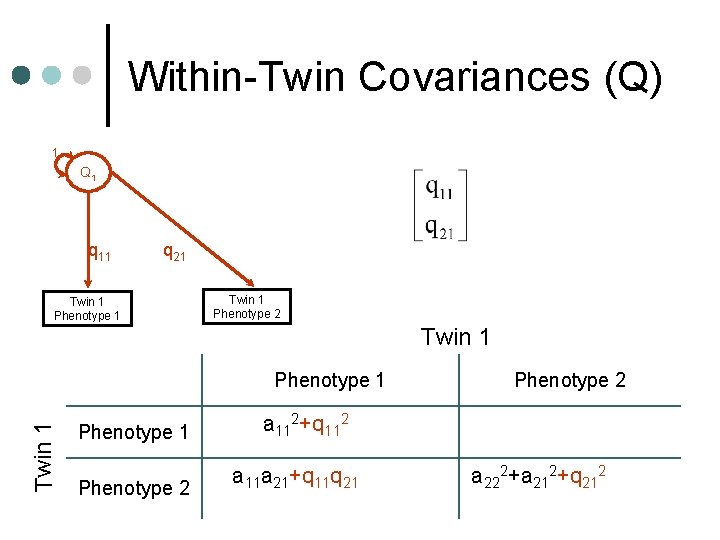
Within-Twin Covariances (Q) 1 Q 1 q 11 q 21 Twin 1 Phenotype 2 Twin 1 Phenotype 1 a 112+q 112+e 112 Phenotype 2 a 11 a 21+q 11 q 21+e 11 e 21 Phenotype 2 a 222+a 212+q 212+e 222+ e 212

Within-Twin Covariances (E) Twin 1 Phenotype 2 Twin 1 Phenotype 1 e 11 1 E 1 e 22 1 E 2 Twin 1 Phenotype 1 a 112+q 112+e 112 Phenotype 2 a 11 a 21+q 11 q 21+e 11 e 21 Phenotype 2 a 222+a 212+q 212+e 222+ e 212

Observed Covariance Matrix Twin 1 Phenotype 2 Twin 2 Phenotype 1 Phenotype 2 Twin 1 Within-twin covariance Phenotype 1 Variance P 1 Phenotype 2 Covariance P 1 -P 2 Variance P 2 Twin 2 Cross-twin covariance Within-twin covariance Phenotype 1 Within-trait P 1 Cross-trait Variance P 1 Phenotype 2 Cross-trait Within-trait P 2 Covariance P 1 -P 2 Variance P 2

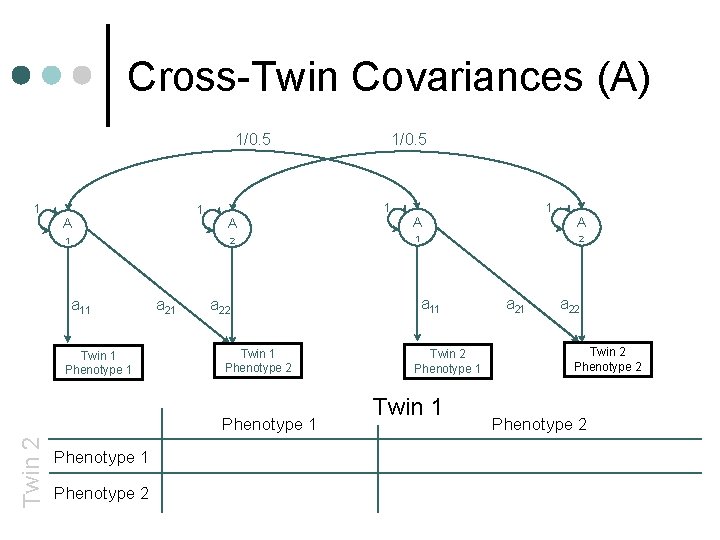
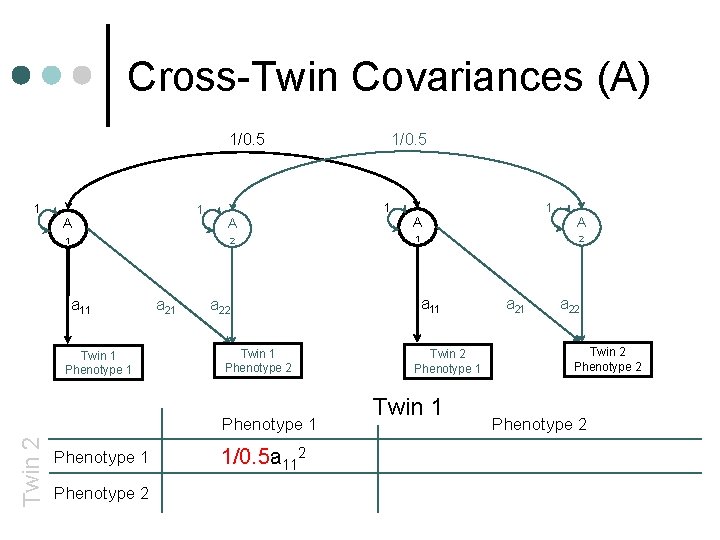
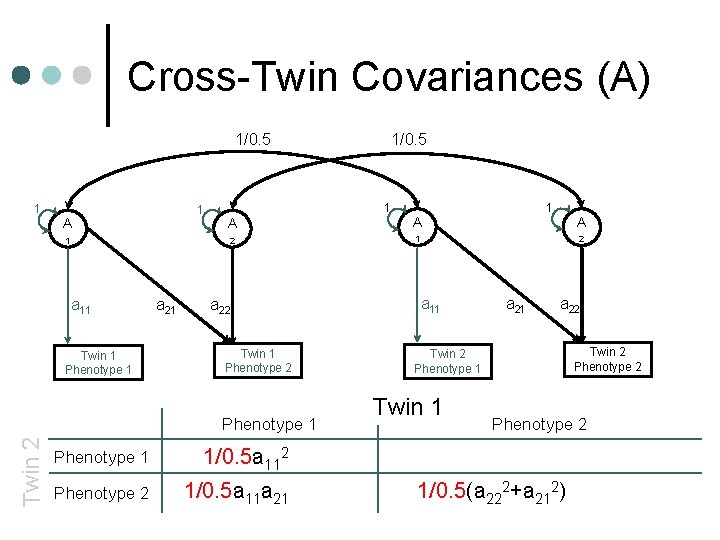
Cross-Twin Covariances (A) 1/0. 5 1 1 A 1 a 11 Twin 1 Phenotype 1 a 21 1 A A 2 1 Twin 1 Phenotype 2 1 A 2 a 11 a 22 Phenotype 1 Twin 2 1/0. 5 Twin 2 Phenotype 1 Twin 1 a 22 Twin 2 Phenotype 1 Phenotype 2 +c 11 c 21 +c 222+c 212

Cross-Twin Covariances (A) 1/0. 5 1 1 A 1 a 11 Twin 1 Phenotype 1 a 21 1 A A 2 1 Twin 1 Phenotype 2 Phenotype 1 1/0. 5 a 112+ Phenotype 2 +c 11 c 21 1 A 2 a 11 a 22 Phenotype 1 Twin 2 1/0. 5 Twin 2 Phenotype 1 Twin 1 a 22 Twin 2 Phenotype 2 +c 222+c 212

Cross-Twin Covariances (A) 1/0. 5 1 1 A 1 a 11 Twin 1 Phenotype 1 a 21 1 A A 2 1 a 22 Twin 1 Phenotype 2 Phenotype 1 Twin 2 1/0. 5 Phenotype 1 1/0. 5 a 112+c 112 Phenotype 2 1/0. 5 a 11 a 21+c 11 c 21 1 A 2 a 11 Twin 2 Phenotype 1 Twin 1 a 22 Twin 2 Phenotype 2 +c 222+c 212

Cross-Twin Covariances (A) 1/0. 5 1 1 A 1 a 11 Twin 1 Phenotype 1 a 21 1 A A 2 1 a 22 Twin 1 Phenotype 2 Phenotype 1 Twin 2 1/0. 5 Phenotype 1 1/0. 5 a 112+c 112 Phenotype 2 1/0. 5 a 11 a 21+c 11 c 21 1 A 2 a 11 Twin 2 Phenotype 1 Twin 1 a 22 Twin 2 Phenotype 2 1/0. 5(a 222+a 212)+c 222+c 212

Cross-Twin Covariances (Q) π 1 1 Q Q 1 1 q 21 q 11 Twin 1 Phenotype 2 Twin 2 Phenotype 1 1/0. 5 a 112+πq 112 Phenotype 2 1/0. 5 a 11 a 21+ π q 11 q 21 Twin 2 Phenotype 2 1/0. 5(a 222+a 212)+ π q 212

Predicted Model Twin 1 Phenotype 2 Twin 2 Phenotype 1 Phenotype 2 Twin 1 Within-twin covariance Phenotype 1 Phenotype 2 a 112+q 112+e 112 a 11 a 21+q 11 q 21+e 11 e 21 a 222+a 212+q 212+ e 222+e 212 Twin 2 Cross-twin covariance Phenotype 1 Phenotype 2 1/. 5 a 112+ π q 112 1/. 5 a 11 a 21+ πq 11 q 21 Within-twin covariance a 112+q 112+e 112 1/. 5(a 222+a 212)+ π q 212 a 11 a 21+q 11 q 21+e 11 e 21 a 222+a 212+q 212+ e 222+e 212

Running MV linkage analysis… ¢ Numerous programs l l Mx Solar Loki Merlin • Repeated measures ¢ ¢ Computationally intensive Multiple boundary issues l p-values difficult to obtain Emu (not e-moo)

MV association… ¢ Maximum likelihood – factor based approach l ¢ Canonical Correlation approach l ¢ Mx Plink Principal components approach l F-bat Funnel Web Spider

Maximum likelihood approach ¢ Unrelated individuals Shared variance due to a common factor l Residual non-shared variance l ¢ Family based data l ACE type models Galah

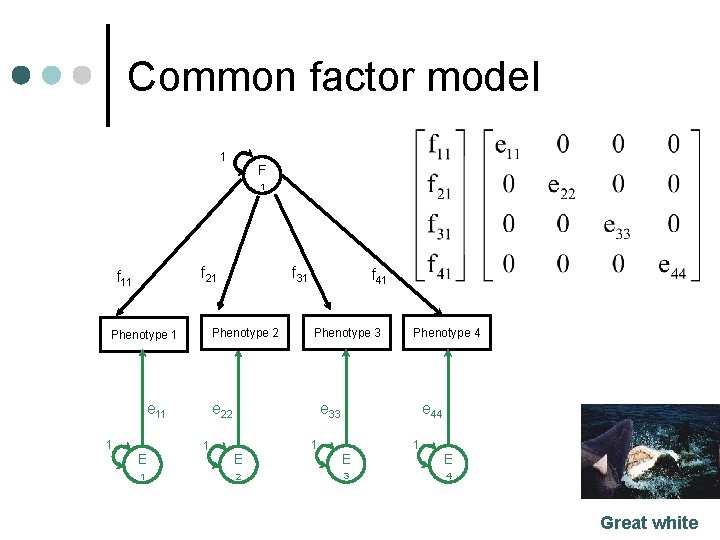
Common factor model 1 F 1 f 21 f 11 Phenotype 2 Phenotype 1 e 11 1 E 1 f 31 f 41 Phenotype 3 e 33 e 22 1 Phenotype 4 E 2 1 e 44 E 3 1 E 4 Great white

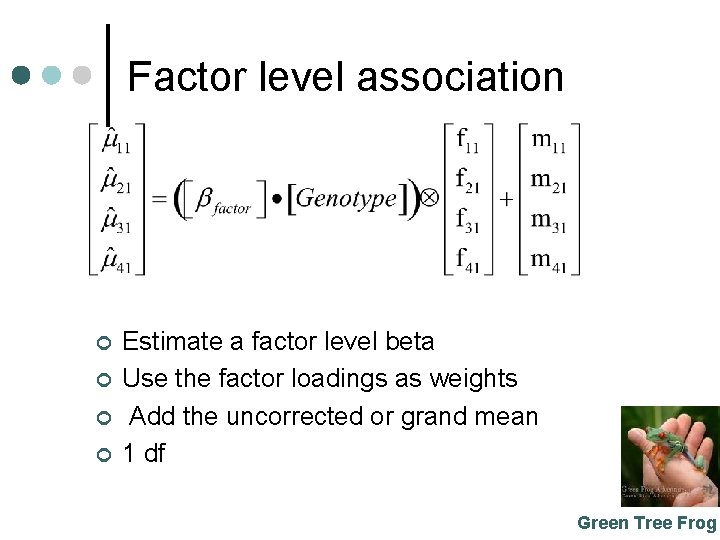
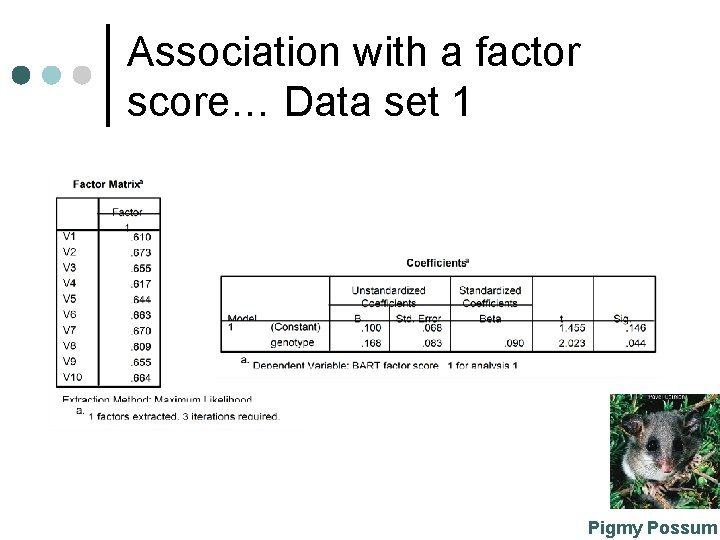
Factor level association ¢ ¢ Estimate a factor level beta Use the factor loadings as weights Add the uncorrected or grand mean 1 df Green Tree Frog

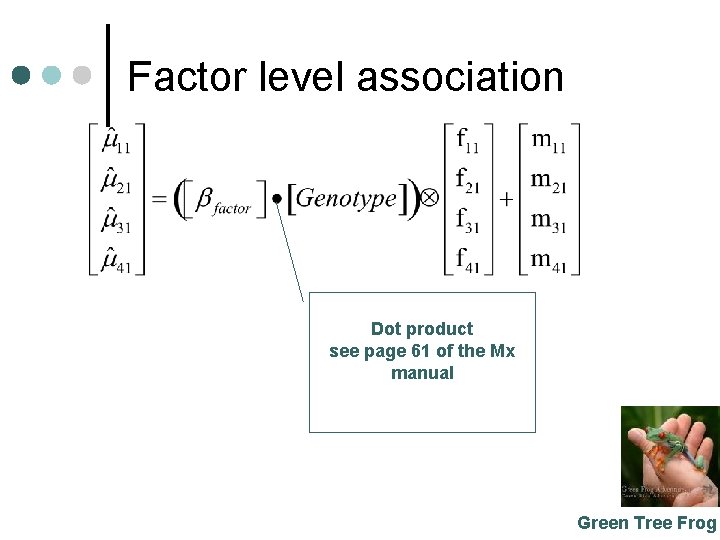
Factor level association Dot product see page 61 of the Mx manual Green Tree Frog

Factor level association Kronecker product see page 61 of the Mx manual Green Tree Frog

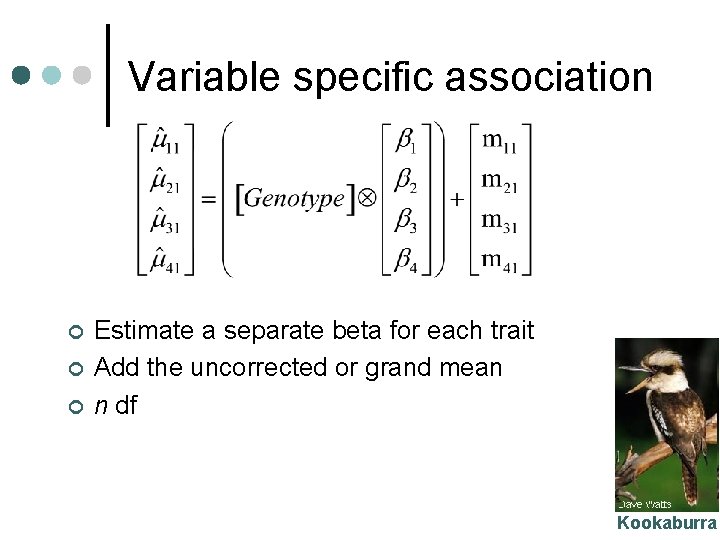
Variable specific association ¢ ¢ ¢ Estimate a separate beta for each trait Add the uncorrected or grand mean n df Kookaburra

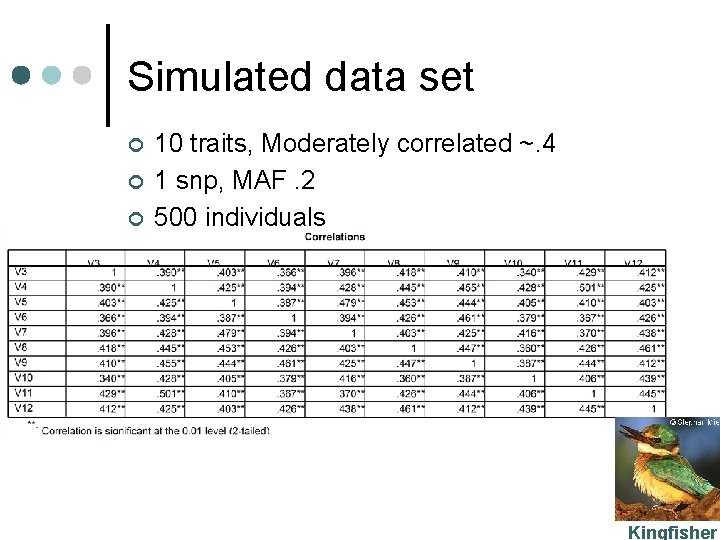
Simulated data set ¢ ¢ ¢ 10 traits, Moderately correlated ~. 4 1 snp, MAF. 2 500 individuals Kingfisher

factorlevel. mx… dataset. ped Example Factor level association script - Boulder 2009 Sarah Medland Data NI=15 NGroups=1 Rec file=dataset. ped Labels fid iid a 1 a 2 genotype V 1 V 2 V 3 V 4 V 5 V 6 V 7 V 8 V 9 V 10 Select genotype V 1 V 2 V 3 V 4 V 5 V 6 V 7 V 8 V 9 V 10 ; Definition genotype ; Begin Matrices; F full 10 1 free R diag 10 10 free M full 10 1 free B full 1 1 free G full 1 1 End Matrices; ! factor ! residuals ! grand means ! association beta ! genotype Numbat

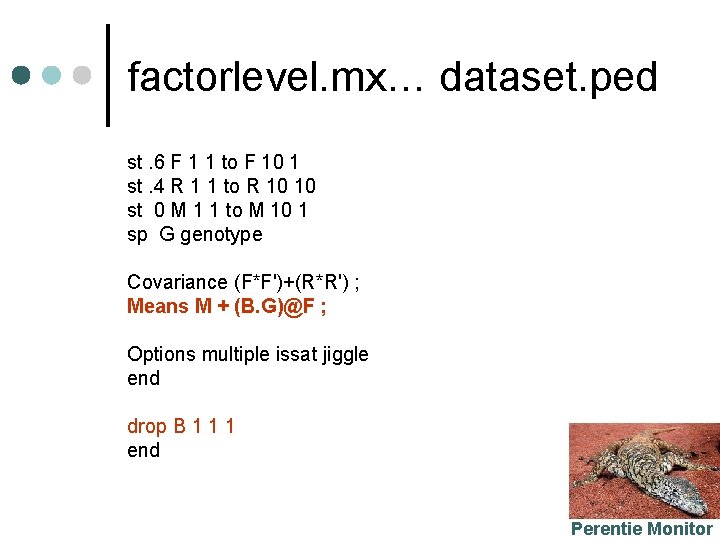
factorlevel. mx… dataset. ped st. 6 F 1 1 to F 10 1 st. 4 R 1 1 to R 10 10 st 0 M 1 1 to M 10 1 sp G genotype Covariance (F*F')+(R*R') ; Means M + (B. G)@F ; Options multiple issat jiggle end drop B 1 1 1 end Perentie Monitor

variablespecific. mx… dataset. ped Example Factor level association script - Boulder 2009 Sarah Medland Data NI=15 NGroups=1 Rec file=dataset. ped Labels fid iid a 1 a 2 genotype V 1 V 2 V 3 V 4 V 5 V 6 V 7 V 8 V 9 V 10 Select genotype V 1 V 2 V 3 V 4 V 5 V 6 V 7 V 8 V 9 V 10 ; Definition genotype ; Begin Matrices; F full 10 1 free R diag 10 10 free M full 10 1 free B full 10 1 free G full 1 1 End Matrices; ! factor ! residuals ! grand means ! association beta ! genotype Quoll

variablespecific. mx… dataset. ped st. 6 F 1 1 to F 10 1 st. 4 R 1 1 to R 10 10 st 0 M 1 1 to M 10 1 sp G genotype Covariance (F*F')+(R*R') ; Means M + (G@B). F ; Options multiple issat jiggle end drop B 1 1 1 to B 1 10 1 end Red back spider

Your task ¢ Run both FL and VS tests in Mx for the first data set Edit the data file name l Calculate the p-value for the VS test using excel… l Which variables are associated? ¢ What is the mean for variable 1 by genotype? ¢

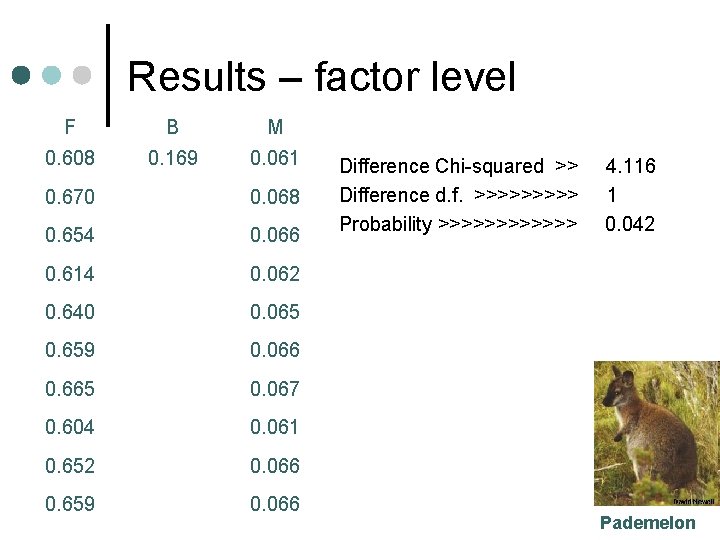
Results – factor level F B M 0. 608 0. 169 0. 061 0. 670 0. 068 0. 654 0. 066 0. 614 0. 062 0. 640 0. 065 0. 659 0. 066 0. 665 0. 067 0. 604 0. 061 0. 652 0. 066 0. 659 0. 066 Difference Chi-squared >> Difference d. f. >>>>> Probability >>>>>> 4. 116 1 0. 042 Pademelon

Association with a factor score… Data set 1 Pigmy Possum

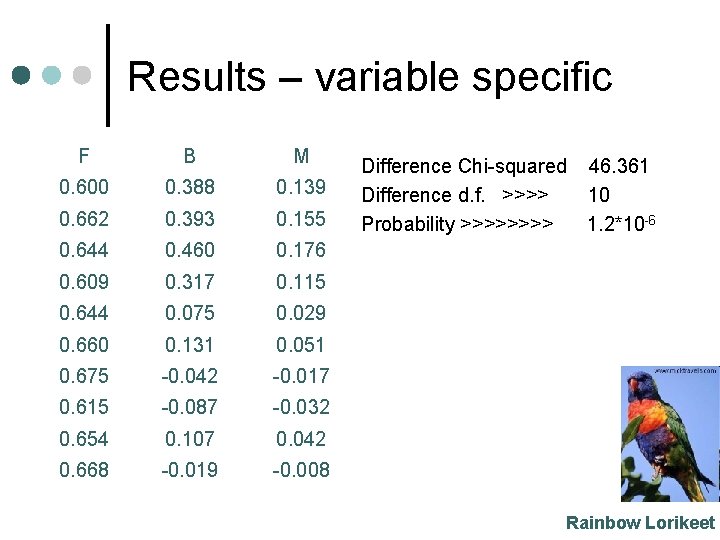
Results – variable specific F B M 0. 600 0. 388 0. 139 0. 662 0. 393 0. 155 0. 644 0. 460 0. 176 0. 609 0. 317 0. 115 0. 644 0. 075 0. 029 0. 660 0. 131 0. 051 0. 675 -0. 042 -0. 017 0. 615 -0. 087 -0. 032 0. 654 0. 107 0. 042 0. 668 -0. 019 -0. 008 Difference Chi-squared Difference d. f. >>>> Probability >>>> 46. 361 10 1. 2*10 -6 Rainbow Lorikeet

Mean under different genotypes -1 AA 0 AB 1 BB -0. 249 0. 139 0. 527 Salt water croc

Advantages to the ML approach ¢ Completely flexible l Can be applied to any model • Longitudinal models • Simplex/Autoregressive processed Easy to add dominance etc l Covariates l Extends to family data l Stone fish

Disadvantages Correction for multiple testing? ¢ FL & VS tests provide complementary information ¢ Inflation of type 1 error ¢ l use a Bonferroni correction if you use both Taipan

Tiger Snake Wombat Tassie Tiger Tree Kangaroo Thorny Devil Zebra finch Tassie Devil
 Sarah medland
Sarah medland Loader design options
Loader design options Multivariate analysis of variance and covariance
Multivariate analysis of variance and covariance Advanced and multivariate statistical methods
Advanced and multivariate statistical methods Differentialgleichung logistisches wachstum
Differentialgleichung logistisches wachstum Multivariate binomial distribution
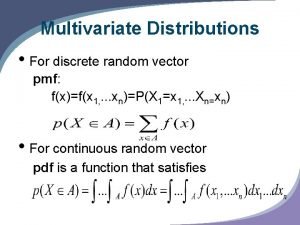
Multivariate binomial distribution Multivariate pdf
Multivariate pdf What is multivariate analysis?
What is multivariate analysis? Multivariate regression spss
Multivariate regression spss Multivariate normal distribution mle
Multivariate normal distribution mle Maximum a posteriori estimation for multivariate gaussian
Maximum a posteriori estimation for multivariate gaussian Multivariate pdf
Multivariate pdf Multivariate vs bivariate
Multivariate vs bivariate Multiple variance analysis
Multiple variance analysis Manova with repeated measures
Manova with repeated measures Ratio test
Ratio test Multivariate methods in machine learning
Multivariate methods in machine learning Nature of multivariate analysis
Nature of multivariate analysis Multivariate verfahren psychologie
Multivariate verfahren psychologie Univariate vs multivariate
Univariate vs multivariate A and b is
A and b is Multivariate descriptive statistics
Multivariate descriptive statistics Multivariate analysis
Multivariate analysis Multivariate statistics for the environmental sciences
Multivariate statistics for the environmental sciences Multivariate statistical analysis
Multivariate statistical analysis Multivariate analysis
Multivariate analysis Multivariate histogram
Multivariate histogram Multivariate analysis
Multivariate analysis Multivariate pattern analysis
Multivariate pattern analysis Sex linked punnett squares
Sex linked punnett squares Sex determination in drosophilla
Sex determination in drosophilla Section 3 gene linkage and polyploidy
Section 3 gene linkage and polyploidy Section 11-3 exploring mendelian genetics answer key
Section 11-3 exploring mendelian genetics answer key Ppst domain 6 explanation
Ppst domain 6 explanation 11-5 linkage and gene maps
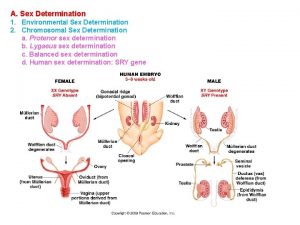
11-5 linkage and gene maps Sex determination and sex linkage
Sex determination and sex linkage Loader and linker
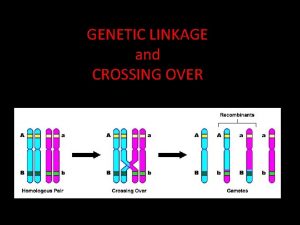
Loader and linker Genetic linkage
Genetic linkage Sexual reproduction and genetics section 1 meiosis
Sexual reproduction and genetics section 1 meiosis Section 11-5 linkage and gene maps answer key
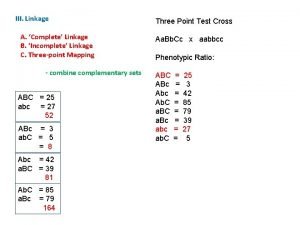
Section 11-5 linkage and gene maps answer key Linkage and recombination
Linkage and recombination Planning and budgeting linkage
Planning and budgeting linkage Genetic linkage and mapping in eukaryotes
Genetic linkage and mapping in eukaryotes Are interest groups linkage institutions
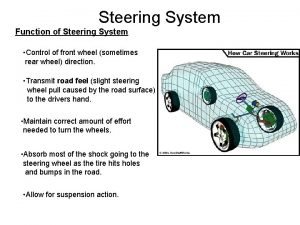
Are interest groups linkage institutions Function of steering wheel
Function of steering wheel 4 wheel
4 wheel Sex linked traits hemophilia punnett square
Sex linked traits hemophilia punnett square Nrlm portal
Nrlm portal Forensic linkage triangle
Forensic linkage triangle Graphical linkage synthesis
Graphical linkage synthesis Media as a linkage institution
Media as a linkage institution Hereditary units
Hereditary units