Extending UCSC Genome Tools Angie Hinrichs UCSC Genome










































- Slides: 57

Extending UCSC Genome Tools Angie Hinrichs UCSC Genome Bioinformatics Group http: //genome. ucsc. edu March 31, 2008

Outline n n UCSC web tools kent/src/ overview n n Adding your own data n n Libraries CGIs (web tools) Command-line utilities What’s already supported Adding code for a new type of track Track metadata Where to learn more

UCSC web tools n n n Genome Browser Table Browser Blat Gene Sorter Genome Graphs … and more

Kent coding in a nutshell n n n Reasonably short functions Defensive checks; die on error (unless server) Consistent naming and indentation Comments at the beginning of each file and function def. ; otherwise only when necessary. Speed! Why import some external lib when Jim can rewrite it better and faster in a day?

kent/src rules of thumb n n n Each program’s source is in a directory with same name as executable Brute-force makefiles (no dependencies beyond. c: . o) Running a command with no arguments shows usage

An aerial tour of kent/src

kent/src important subdirs n n n inc, lib, jk. Own. Lib utils, blat, parasol, and early work hg n n n inc, lib hg* make. Db utils mouse. Stuff, rat. Stuff, …

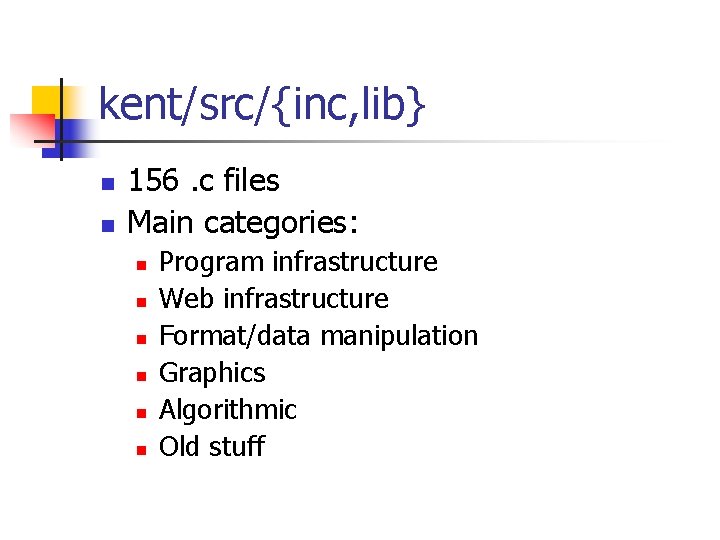
kent/src/{inc, lib} n n 156. c files Main categories: n n n Program infrastructure Web infrastructure Format/data manipulation Graphics Algorithmic Old stuff

src/lib: program infrastructure n n n options. c errabort. c, err. Catch. c verbose. c n n n n common. c dystring. c linefile. c n n n memalloc. c obscure. c portimpl. c localmem. c pipeline. c file. Path. c log. c

src/lib: web infrastructure n n cheapcgi. c htmshell. c internet. c n n n apache. Log. c html. Page. c qa. c

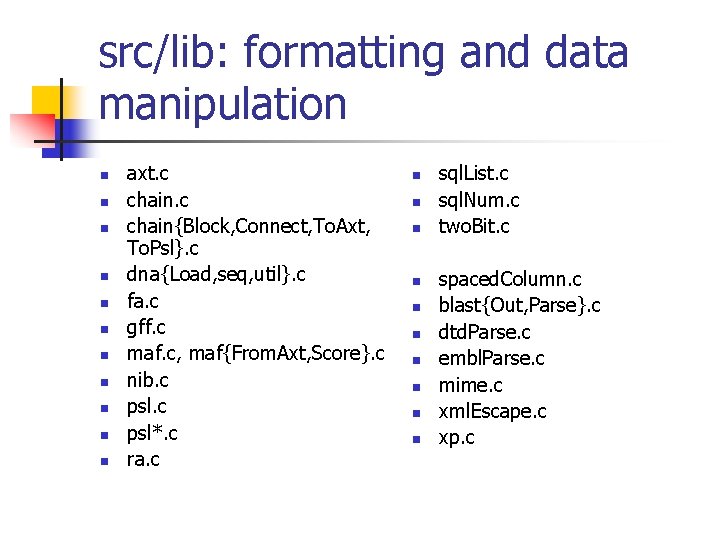
src/lib: formatting and data manipulation n n axt. c chain{Block, Connect, To. Axt, To. Psl}. c dna{Load, seq, util}. c fa. c gff. c maf. c, maf{From. Axt, Score}. c nib. c psl*. c ra. c n n n n n sql. List. c sql. Num. c two. Bit. c spaced. Column. c blast{Out, Parse}. c dtd. Parse. c embl. Parse. c mime. c xml. Escape. c xp. c

src/lib: Graphics n n n n gfx. Poly. c gif*. c memgfx. c mg*. c ps{Gfx, Poly}. c v. Gfx. c v. Gif. c

src/lib: Algorithmic n n hash. c bits. c wildcmp. c sub. Text. c n n n bin. Range. c space. Saver. c trix. c axt. Affine. c rb. Tree. c phylo. Tree. c range. Tree. c n n dlist. c di. Graph. c quick. Heap. c correlate. c kx. Tok. c tokenizer. c box. Clump. c, box. Lump. c, dna. Markov. c, dna. Motif. c, histogram. c, int. Exp. c, int. Val. Tree. c, keys. c, md 5. c, pair. Hmm. c, spaced. Seed. c, …

Linked lists are everywhere n Struct definition convention: struct foo /* A blah. */ { struct foo *next; char *name; … n sl*, sl. Name* routines in src/lib/common. c

src/lib/common. c n n n n String copying and manipulation Bounds-checking versions of sprintf, strcpy, strcat File I/O with error checks byte/ int / double array manipulation Sort, reverse, count, cat, shift/pop, add, remove from generic lists Generic named-item lists: above plus search, to/from strings w/separators, mem alloc and freeing Generic container lists Name-value pair lists

kent/src/hg/{inc, lib} n n 412. c files Main categories: n n Infrastructure for our CGIs + database auto. Sql-generated database table interfaces (449. as files, 545. sql files) More file formats, shared util code Old stuff

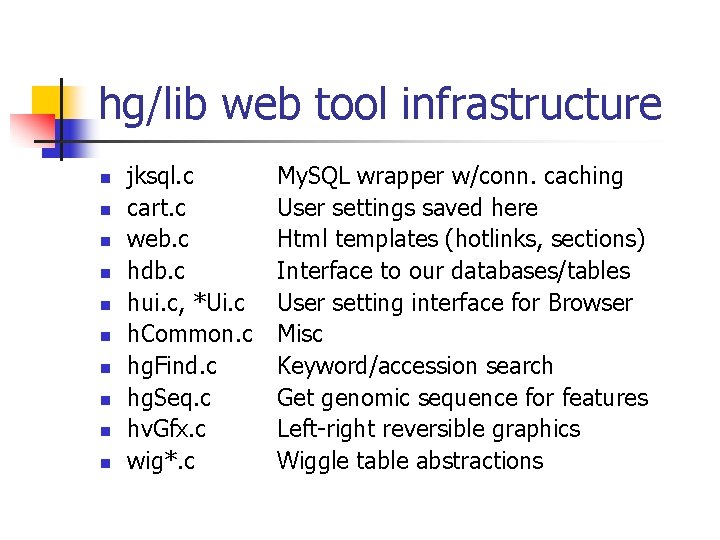
hg/lib web tool infrastructure n n n n n jksql. c cart. c web. c hdb. c hui. c, *Ui. c h. Common. c hg. Find. c hg. Seq. c hv. Gfx. c wig*. c My. SQL wrapper w/conn. caching User settings saved here Html templates (hotlinks, sections) Interface to our databases/tables User setting interface for Browser Misc Keyword/accession search Get genomic sequence for features Left-right reversible graphics Wiggle table abstractions

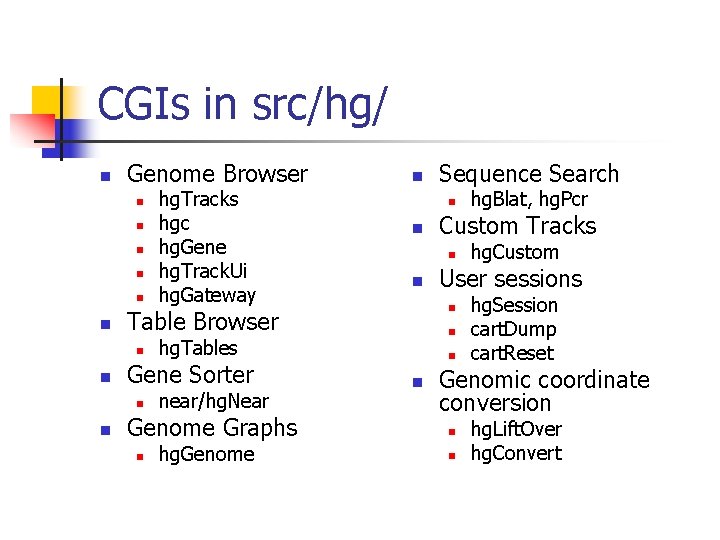
CGIs in src/hg/ n Genome Browser n n near/hg. Near Genome Graphs n hg. Genome hg. Custom User sessions n n hg. Blat, hg. Pcr Custom Tracks n hg. Tables Gene Sorter Sequence Search n Table Browser n n hg. Tracks hgc hg. Gene hg. Track. Ui hg. Gateway n hg. Session cart. Dump cart. Reset Genomic coordinate conversion n n hg. Lift. Over hg. Convert

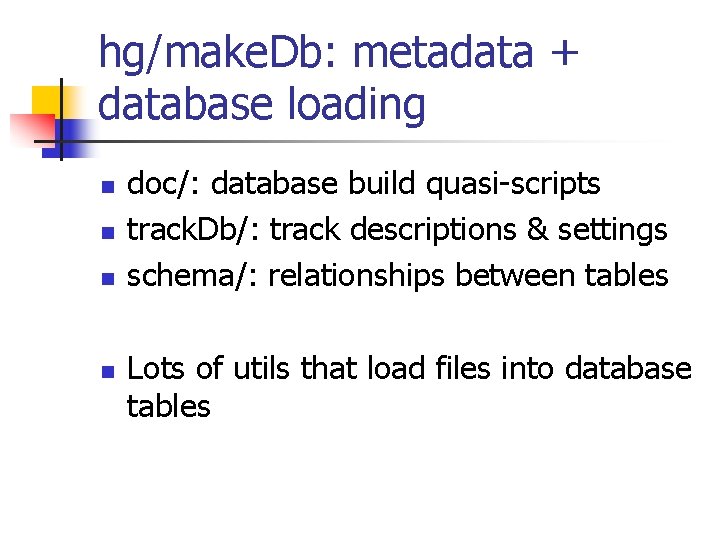
hg/make. Db: metadata + database loading n n doc/: database build quasi-scripts track. Db/: track descriptions & settings schema/: relationships between tables Lots of utils that load files into database tables

Command line utilities n n n Alignment Filtering, mapping, & other transforms Database loading Format conversion Database-building automation scripts new. Prog

Command line conventions n n n Run program w/o arguments usage Filename args can be stdin or stdout Append = to argument to specify value (e. g. my. Prog –iter=10 input. File) n db. Prog db table file 1 [file 2 […]] n Output may appear first or last

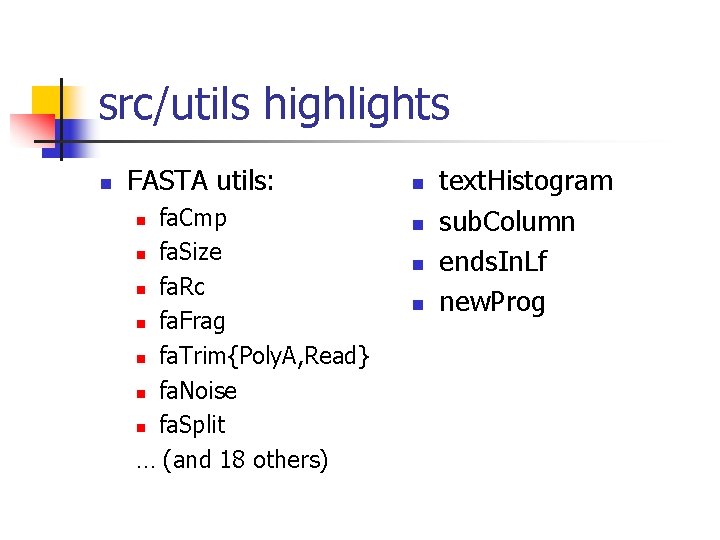
src/utils highlights n FASTA utils: fa. Cmp n fa. Size n fa. Rc n fa. Frag n fa. Trim{Poly. A, Read} n fa. Noise n fa. Split … (and 18 others) n n n text. Histogram sub. Column ends. In. Lf new. Prog

Example util: src/utils/fa. Some. Records n n n option. Specs / option. Init line. File. Open, line. File. Row, line. File. Next hash. New, hash. Add, hash. Lookup clone. String, next. Word, free. Mem must. Open

Utils in src/hg/ n n Hundreds: 43 psl*, many gene. Pred*, agp*, bed* Other notables: n n n lift. Up, lift. Over, ortho. Map, psl. Map hgsql*, hg. Get. Ann, hg. Select xml. To. Sql, auto{Sql, Dtd, Xml} hg. Blat. Test, hg. Tables. Test, qa/* check. Table. Coords, check. Hg. Find. Spec

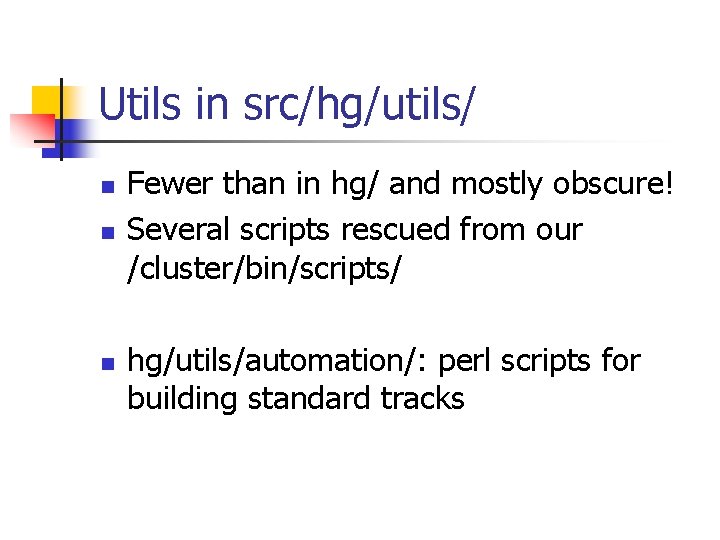
Utils in src/hg/utils/ n n n Fewer than in hg/ and mostly obscure! Several scripts rescued from our /cluster/bin/scripts/ hg/utils/automation/: perl scripts for building standard tracks

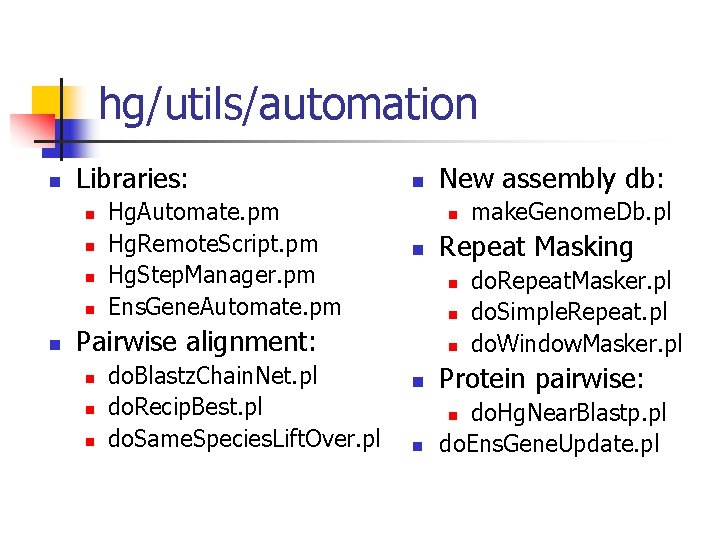
hg/utils/automation n Libraries: n n n Hg. Automate. pm Hg. Remote. Script. pm Hg. Step. Manager. pm Ens. Gene. Automate. pm n n n do. Blastz. Chain. Net. pl do. Recip. Best. pl do. Same. Species. Lift. Over. pl make. Genome. Db. pl Repeat Masking n n Pairwise alignment: n New assembly db: n do. Repeat. Masker. pl do. Simple. Repeat. pl do. Window. Masker. pl n Protein pairwise: n do. Hg. Near. Blastp. pl do. Ens. Gene. Update. pl n

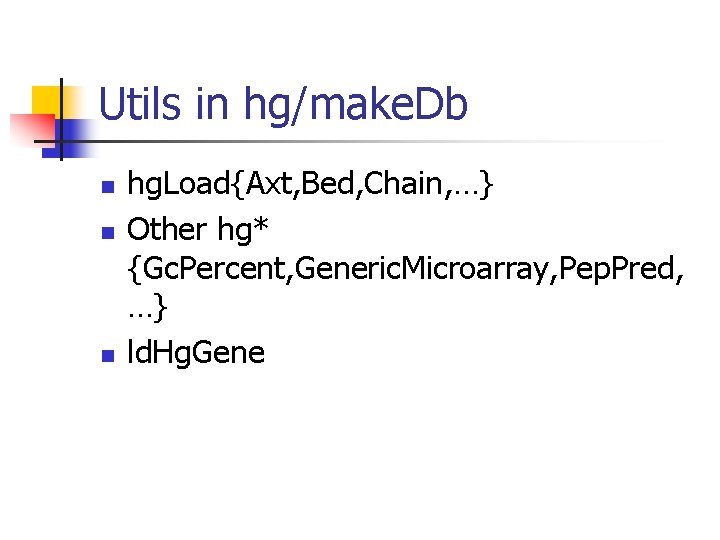
Utils in hg/make. Db n n n hg. Load{Axt, Bed, Chain, …} Other hg* {Gc. Percent, Generic. Microarray, Pep. Pred, …} ld. Hg. Gene

OK, so where are the chain and net utils? n n hg/mouse. Stuff: axt*, chain*, lav*, maf*, net* hg/rat. Stuff: more maf*

Adding your own data

Data processing tips n n n Use examples from make. Db/*. txt Document! (as in make. Db/doc/*. txt) Know your shell, sed, awk, perl (join, cut, …) Most programs can read. gz files Use pipes to avoid unnecessary file I/O

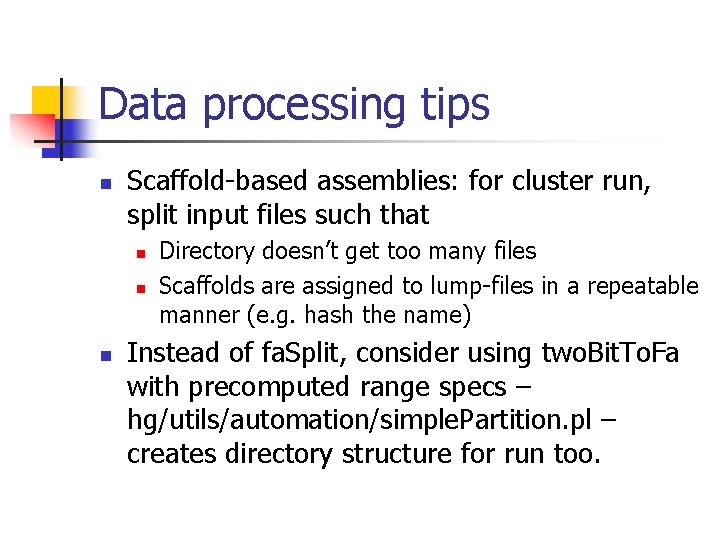
Data processing tips n Scaffold-based assemblies: for cluster run, split input files such that n n n Directory doesn’t get too many files Scaffolds are assigned to lump-files in a repeatable manner (e. g. hash the name) Instead of fa. Split, consider using two. Bit. To. Fa with precomputed range specs – hg/utils/automation/simple. Partition. pl – creates directory structure for run too.

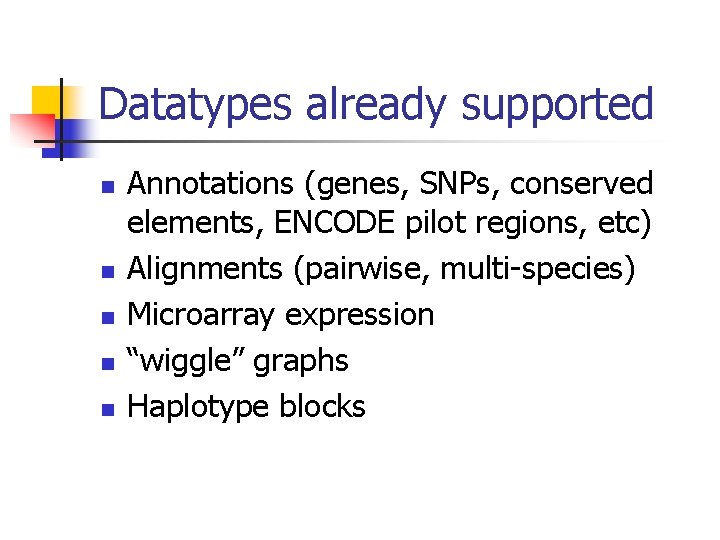
Datatypes already supported n n n Annotations (genes, SNPs, conserved elements, ENCODE pilot regions, etc) Alignments (pairwise, multi-species) Microarray expression “wiggle” graphs Haplotype blocks

Adding a new track? Don’t reinvent the wheel… 1. 2. 3. 4. Temporary? Use custom-track if possible. Use existing data type if possible Use BED (first N applicable fields) Extra info used only on details page? BED and auxiliary table; otherwise, extended BED.

Genome Browser anatomy n hg. Tracks: graphical display n n n n struct track: quasi-object coded in C 19 methods, e. g. load. Items, draw. Items, … Typical “track handler” overrides a few methods, hooks into register. Handler() or fill. In. From. Type() hgc: item details hg. Track. Ui: controls track. Db settings specify defaults cart variables contain user choices

hg. Tracks example: net. Track. c n Punchline at end of file: void net. Methods(struct track *tg) /* Make track group for chain/net alignment. */ { tg->load. Items = net. Load; tg->free. Items = net. Free; tg->draw. Items = net. Draw; tg->item. Name = net. Name; tg->map. Item. Name = net. Name; tg->total. Height = tg. Fixed. Total. Height. No. Overflow; tg->item. Height = tg. Fixed. Item. Height; tg->item. Start = tg. Item. No. Start; tg->item. End = tg. Item. No. End; tg->maps. Self = TRUE; }

net. Track hooks in hg. Tracks. c void fill. In. From. Type(struct track *track, struct track. Db *tdb) /* Fill in various function pointers in track from type field of tdb. */ { … else if (same. Word(type, "net. Align")) { net. Methods(track); } …

The other kind of hg. Tracks. c hook struct track *get. Track. List( struct group **p. Group. List, int vis) … register. Track. Handler("cyto. Band. Ideo", cyto. Band. Ideo. Methods); register. Track. Handler("bac. End. Pairs", bac. End. Pairs. Methods); register. Track. Handler("bac. End. Pairs. Bad", bac. End. Pairs. Bad. Methods); …

Net track details in hgc. c void generic. Click. Handler. Plus( struct track. Db *tdb, char *item. For. Url, char *plus) /* Put up generic track info, with additional text appended after item. */ { … else if (same. String(type, "net. Align")) { if (word. Count < 3) err. Abort("Missing field in net. Align track type field"); generic. Net. Click(conn, tdb, item, start, words[1], words[2]); } …

The other kind of hgc. c hook void do. Middle() /* Generate body of HTML. */ { … else if (same. Word(track, "waba. Cbr")) { do. Hg. Cbr(tdb, item); } else if (starts. With("rmsk", track)) { do. Hg. Repeat(tdb, item); } …

hg. Track. Ui. c: both types of hook in one place void /* { char … else specific. Ui(struct track. Db *tdb) Draw track specific parts of UI. */ *track = tdb->table. Name; if (same. String(track, "fish. Clones")) fish. Clones. Ui(tdb); else if (same. String(track, "recomb. Rate")) recomb. Rate. Ui(tdb); … else if (starts. With("bed. Graph", tdb->type)) wig. Ui(tdb); …

track. Db: a table of track info n n n track. Db table is compiled from track. Db. ra files Main fields: table. Name, short. Label, long. Label, visibility, priority, group, type Extensible: settings field allows arbitrary var value mappings.

track. Db. ra files n n See also make. Db/track. Db/README Up to 3 track. Db. ra files per db. hg 18: n n n make. Db/track. Db. ra make. Db/track. Db/human/track. Db. ra make. Db/track. Db/human/hg 18/track. Db. ra track. html file for track description page New database must be added to track. Db/makefile.

track. Db. ra example track known. Gene short. Label Known Genes long. Label Known Genes Based on SWISS-PROT, Tr. EMBL, m. RNA, and Ref. Seq group genes priority 34 visibility pack color 12, 120 type gene. Pred known. Gene. Pep known. Gene. Mrna id. Xref kg. Alias kg. ID alias hg. Gene on hgsid on direct. Url /cgibin/hg. Gene? hgg_gene=%s&hgg_chrom=%s&hgg_start=%d&hgg_end=%d&hgg_t ype=%s&db=%s base. Color. Use. Cds given base. Color. Default genomic. Codons

Composite tracks track most. Conserved 28 way composite. Track on short. Label Most Conserved long. Label Phast. Conserved Elements, 28 -way Vertebrate Multiz Alignment group comp. Geno priority 103. 4 visibility hide exon. Arrows off show. Top. Scorers 200 type bed 5. track phast. Cons. Elements 28 way. Plac. Mammal sub. Track most. Conserved 28 way short. Label Mammal long. Label Phast. Cons Placental Mammal Conserved Elements, 28 -way Multiz Alignment color 100, 50, 170 priority 1 track phast. Cons. Elements 28 way sub. Track most. Conserved 28 way short. Label Vertebrate long. Label Phast. Cons Vertebrate Conserved Elements, 28 -way Multiz Alignment color 170, 100, 50 priority 2

Adding item position search n Specify search in track. Db. ra http: //genome. ucsc. edu/admin/hg. Find. Spec. How. To. html n Example: search for Affy SNPs: search. Name affy 250 Sty search. Table snp. Array. Affy 250 Sty search. Method exact search. Type bed term. Regex (SNP_A-[0 -9]+) search. Priority 12 padding 250

If you must add a new datatype… n n Consider BED 3+ (genomic position plus other stuff) for baseline Genome and Table Browser support. SQL table design: n n Not too relational – one big old table Use auto. Sql Add bin column (indexing speedup) to. sql Add indexes: (chrom, bin) and maybe name

auto. Sql: c+sql code generator n n n n hg/auto. Sql/ hg/lib/*. as: auto. Sql table specs auto. Sql foo. as foo. {c, h, sql}. sql doctoring: add bin row, add index Move. h to. . /inc. c has functions to read from file, read from db, write to file If you won’t use the. c/. h, don’t check them in!

Using files in addition to database tables n Need to keep raw data in files? Use seq/ext. File tables (see hg. Load. Seq, hg. Load. Maf)

. ra metadata – not just for track. Db n n Gene Sorter has own metadata hg/near/hg. Near. Data/*. ra name go. Similarity short. Label GO Similarity long. Label Number of Shared Gene Ontology Terms type association go. goa. Part priority 7. 1 prot. Key on query. One select go. Id from go. goa. Part where db. Object. Symbol='%s' query. All select db. Object. Symbol, go. Id from go. goa. Part

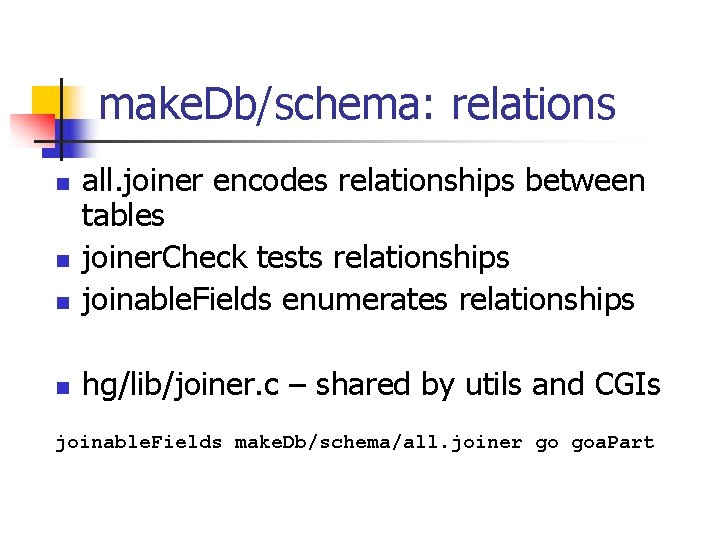
make. Db/schema: relations n all. joiner encodes relationships between tables joiner. Check tests relationships joinable. Fields enumerates relationships n hg/lib/joiner. c – shared by utils and CGIs n n joinable. Fields make. Db/schema/all. joiner go goa. Part

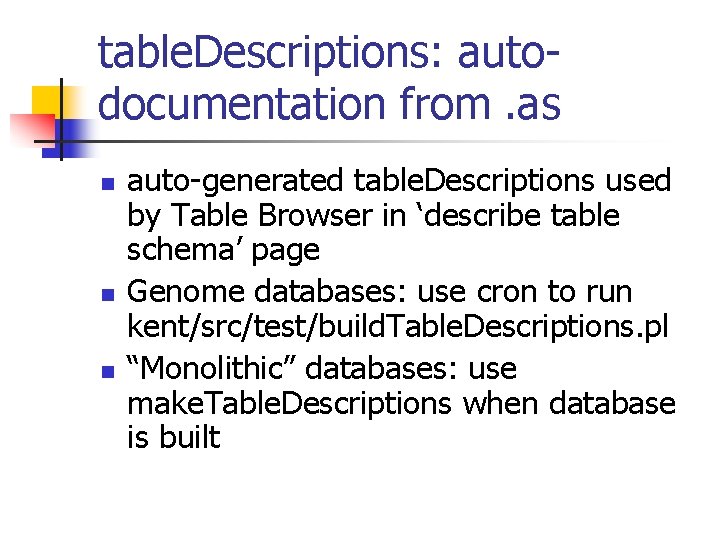
table. Descriptions: autodocumentation from. as n n n auto-generated table. Descriptions used by Table Browser in ‘describe table schema’ page Genome databases: use cron to run kent/src/test/build. Table. Descriptions. pl “Monolithic” databases: use make. Table. Descriptions when database is built

Debugging tips n Run CGIs on command line with settings from URL: cd kent/src/hg/hgc/ /usr/local/apache/cgi-bin-$USER/hgc ‘hgsid=283532&l=23432&r=44879&…’ n n Make. hg. conf (chmod 600) or set HG_CONF to …/cgi-bin-$USER/hg. conf. . /trash (kent/src/hg/trash): symbolic link /usr/local/apache/trash HTTP_COOKIE: set to cookie string displayed at end of cart. Dump if necessary (usually not) Use gdb!

gdb basics … 7 little commands get you so far! n Crashing: gdb /usr/local/apache/cgi-bin-$USER/hgc break err. Abort break sql. Abort run ‘hgsid=283532&l=23432&r=44879&…’ where n Misbehaving: gdb /usr/local/apache/cgi-bin-$USER/hgc break do. My. Track run ‘hgsid=283532&l=23432&r=44879&…’ n print xyz s print sl. Count(that. List) break hgc. c: 3487 c

More info inside kent/src n kent/src/product/README. * n n n README. building. source README. mysql. setup README. track. Db README. debug More mirror-specific README. * kent/src/hg/doc/add. Track. txt

More info by email n n n Send code/data questions to genome-mirror@soe. ucsc. edu Send browser usage questions to genome@soe. ucsc. edu Want more email? Sign up! n n n Can join genome to see lots of Q&A http: //www. soe. ucsc. edu/mailman/listinfo/genome-mirror: Q&A about mirror issues, heads-up about big data genome-announce: major announcements (downtime, release of new db or tool)

More info n n n Search genome@soe. ucsc. edu archives http: //genome. ucsc. edu/FAQ/ Random articles by staff and powerusers: http: //genomewiki. ucsc. edu Good free usage tutorials by Open. Helix http: //www. openhelix. com/

Thanks n UCSC Genome Bioinformatics Group n Funding: NHGRI, HHMI, NCI
 Semi-global alignment
Semi-global alignment Ucsc genome browser tutorial
Ucsc genome browser tutorial Paul hinrichs
Paul hinrichs Stefan hinrichs
Stefan hinrichs Angie loved to draw theme
Angie loved to draw theme Angie becker kudelka
Angie becker kudelka Jenny hated reading class theme
Jenny hated reading class theme Angie whittington
Angie whittington Angie rupert
Angie rupert Angie fowble
Angie fowble Usenet message angie
Usenet message angie The land of gallon
The land of gallon Angie bush
Angie bush When was angie thomas born
When was angie thomas born Meaning
Meaning Theme meaning and examples
Theme meaning and examples Angie s. ist in fahrt
Angie s. ist in fahrt Angie french
Angie french Parker hannifin cyberattack
Parker hannifin cyberattack Angie varona sexting
Angie varona sexting Angie stone weight loss
Angie stone weight loss Powerschool waukee
Powerschool waukee Angie dirty feet
Angie dirty feet Jenny puchovier was so excited theme
Jenny puchovier was so excited theme Talker doer supporter controller test
Talker doer supporter controller test Angie loved to draw theme
Angie loved to draw theme Angie travel
Angie travel Angie corinne
Angie corinne Aaron dugmore
Aaron dugmore Angie bennett
Angie bennett Angie van eyck
Angie van eyck What is a theme
What is a theme Chapter 7 extending mendelian genetics vocabulary practice
Chapter 7 extending mendelian genetics vocabulary practice Holandric genes
Holandric genes Xbar theory
Xbar theory Large land mass
Large land mass Extending x-bar theory
Extending x-bar theory Chapter 7 extending mendelian genetics study guide answers
Chapter 7 extending mendelian genetics study guide answers Extending mendelian genetics
Extending mendelian genetics Emerging proficient extending
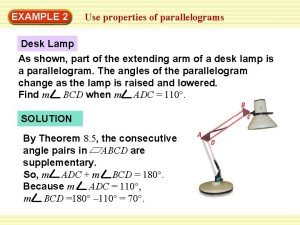
Emerging proficient extending Parallelogram lamp
Parallelogram lamp X-bar schema
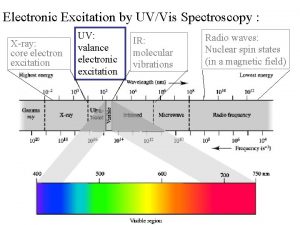
X-bar schema Spectrum xray
Spectrum xray A high point of land extending into water
A high point of land extending into water Extending surface area and volume
Extending surface area and volume Carrier female genotype
Carrier female genotype Uv spectra of dienes
Uv spectra of dienes Problem 8-3 extending amounts across the work sheet
Problem 8-3 extending amounts across the work sheet Identify the landform
Identify the landform Extending oblivious transfers efficiently
Extending oblivious transfers efficiently Marking tools in sewing
Marking tools in sewing Chapter 14 the human genome making karyotypes answer key
Chapter 14 the human genome making karyotypes answer key Savant genome browser
Savant genome browser Section 14-3 human molecular genetics answers
Section 14-3 human molecular genetics answers Mash genome
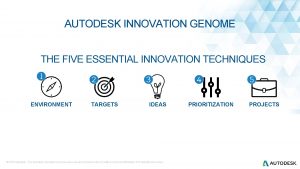
Mash genome Innovation genome project
Innovation genome project Open reading frame
Open reading frame Genome
Genome