Exploring Protein Sequences Part 2 Part 1 Part







- Slides: 29

Exploring Protein Sequences - Part 2 Part 1: Part 2: Patterns and Motifs Profiles Hydropathy Plots Transmembrane helices Antigenic Prediction Signal Peptides Repeats Coiled Coils Linkers Protein Domains Domain databases Celia van Gelder CMBI Radboud University December 2005 ©CMBI 2005

Definition of protein domains • Group of residues with high contact density, number of contacts within domains is higher than the number of contacts between domains. • A stable unit of protein structure that can fold autonomously • A rigid body linked to other domains by flexible linkers • A portion of the protein that can be active on its own if you remove it from the rest of the protein. ©CMBI 2005

Protein Domains • Domains can be 25 to 500 residues long; most are less than 200 residues • The average protein contains 2 or 3 domains • The total number of different types of domains ~1000 – 3000 • The same or similar domains are found in different proteins. “Nature is a ‘tinkerer’ and not an inventor” (Jacob, 1977). “Nature is smart but lazy” • Usually, each domain plays a specific role in the function of the protein. ©CMBI 2005

Linkers Domain linkers link the protein domains together and have been found to contain an amino acid signature that is distinct from the structurally compact domains. Average linker size 8 -9 amino acids Linkers are susceptible for protease attack and they are flexible. ©CMBI 2005

Protein Domain Databases Even though the structure of a domain is not always known it is still possible to define the domain boundaries from sequence alone Many of the common domains have already been defined in domain databases Advantages: • Pre-annotated domains • Easy interpretation of domain structure Problem: • Not trivial to define domain boundaries unambiguously ©CMBI 2005

Protein Domains http: //ip 30. eti. uva. nl/ember-demo/ch 3

Domain databases (2) Generation #entries Pfam. A manual 7503 families Pfam. B automatic >140, 000 families Prints manual 11, 170 motifs Prosite Profiles manual 577 profiles Blocks automatic 28, 337 blocks, 5733 groups SMART manual 667 HMMs Pro. Dom automatic 501, 917 domain families ©CMBI 2005

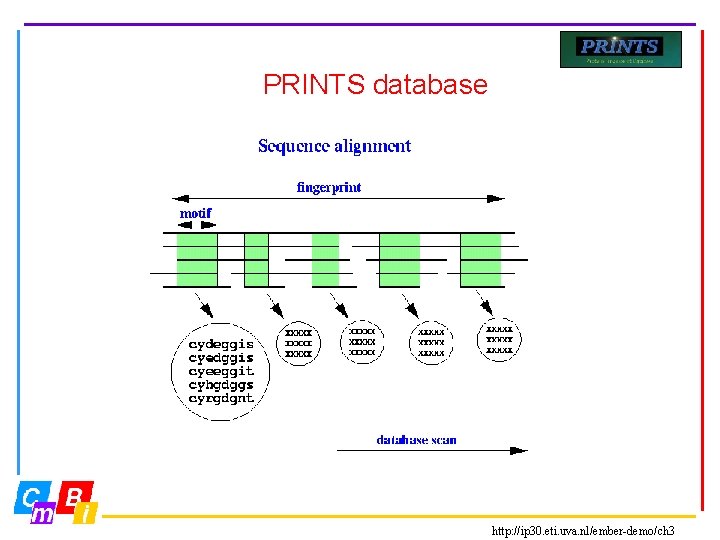
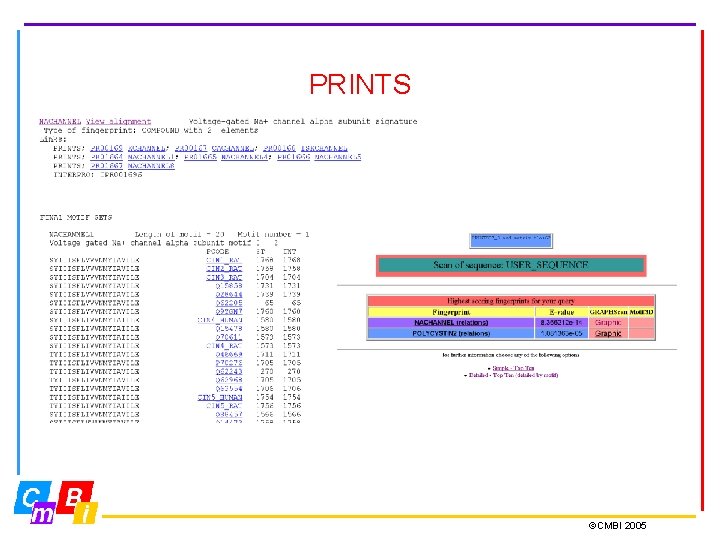
PRINTS database • Most protein families are characterised not by one, but by several conserved motifs • Fingerprints are groups of conserved motifs excised from sequence alignments • Taken together, they provide diagnostic family signatures. They are the basis of the PRINTS database, and are stored in the form of aligned motifs • Input about protein families is done manually • True members match all elements of the fingerprint in order, subfamily members may match part of fingerprint ©CMBI 2005

PRINTS database http: //ip 30. eti. uva. nl/ember-demo/ch 3

PRINTS ©CMBI 2005

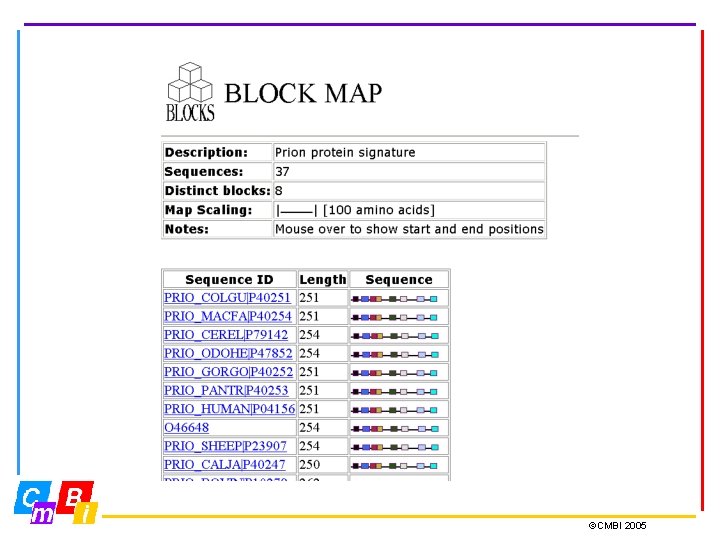
BLOCKS database Blocks are multiply aligned ungapped segments corresponding to the most highly conserved regions of proteins. The blocks for the BLOCKs database are made automatically by looking for the most highly conserved regions in groups of proteins documented in Inter. Pro. Version 14. 1 of the BLOCKS Database consists of 28, 337 blocks representing 5733 groups documented in Inter. Pro 8. 1 (february 2005) To ensure complete coverage it is recommended that both the PRINTS and the BLOCKS database be searched ©CMBI 2005

©CMBI 2005

©CMBI 2005

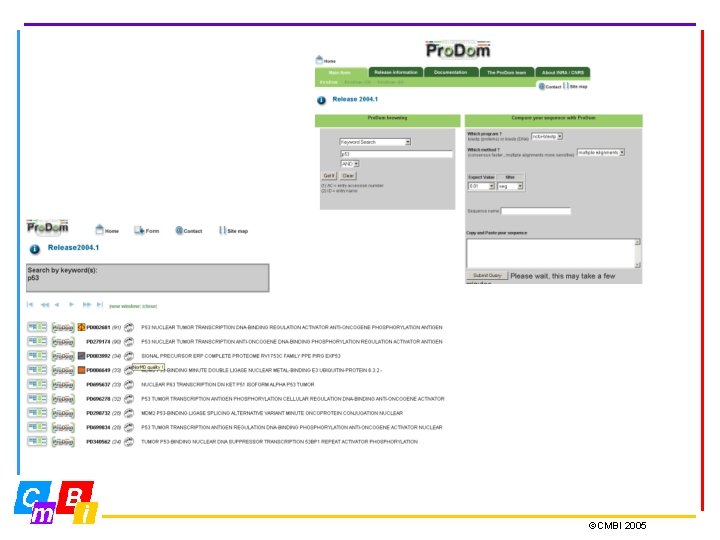
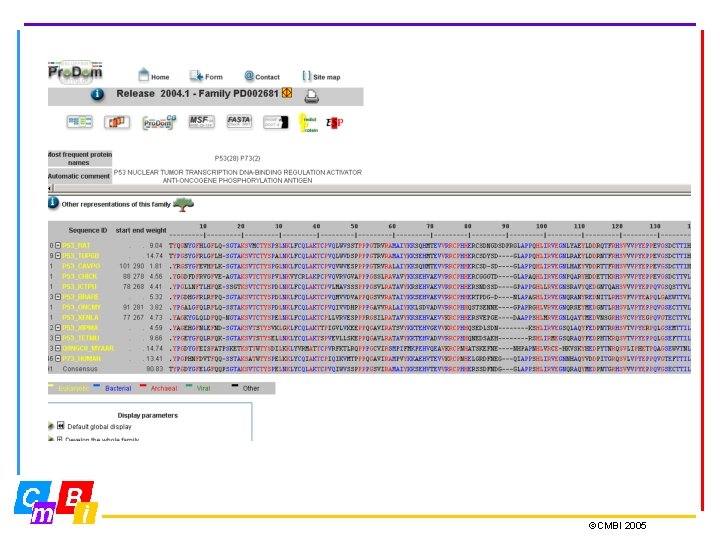
Pro. Dom: The Protein Domain Database • Pro. Dom is a comprehensive set of protein domain families automatically generated • Each entry provides a multiple sequence alignment of homologous domains and a family consensus sequence. • Current Pro. Dom release: Pro. Dom 2004. 1, June 2004, 501917 domain families ©CMBI 2005

©CMBI 2005

©CMBI 2005

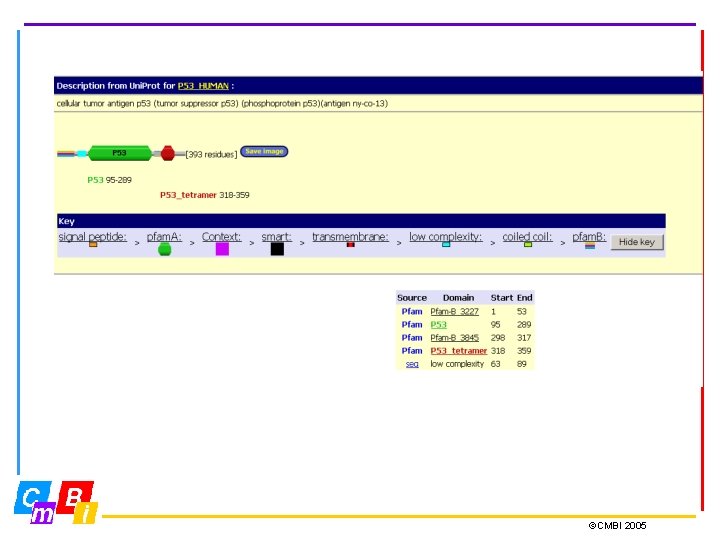
Pfam (Protein families) is a large collection of multiple sequence alignments and hidden Markov models covering many common protein domains and families. For each family in Pfam you can: • Look at multiple alignments • View the domain organisation of proteins • Examine species distribution • Follow links to other databases • View known protein structures ©CMBI 2005

Pfam Two distinct parts: –Pfam-A entries are manually curated 7503 families –Pfam-B entries automatically generated clusters >140, 000 (not covered by Pfam-A) New: i. Pfam is a resource that describes domain-domain interactions that are observed in known structures ©CMBI 2005

©CMBI 2005

©CMBI 2005

SMART - Simple Modular Architecture Research Tool Domain families found in: 1) signalling 2) nuclear 3) extracellular 4) other Current version 5. 0: Number of SMART HMMs: 669 You can use SMART in two different modes: normal or genomic. ©CMBI 2005

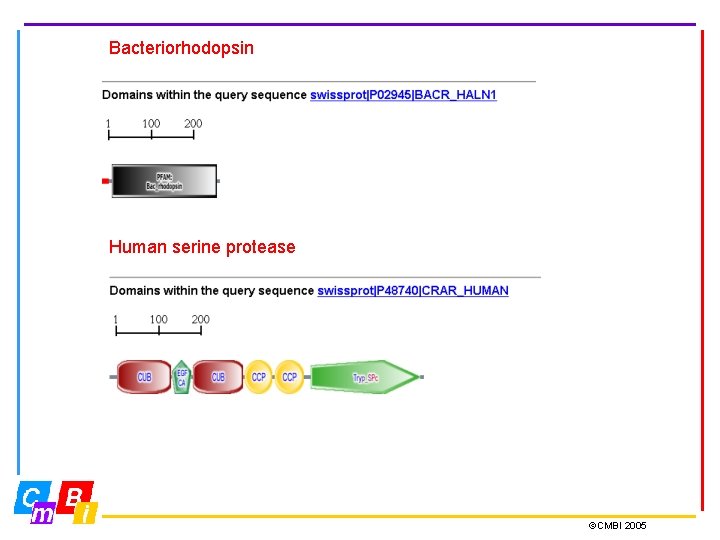
Bacteriorhodopsin Human serine protease ©CMBI 2005

Limitations of domain databases • Patterns not present for all families of proteins • Multiple sequence alignment to define patterns could be inaccurate due to an automatic alignment • Low number of sequences from different species could result in inaccurate patterns ©CMBI 2005

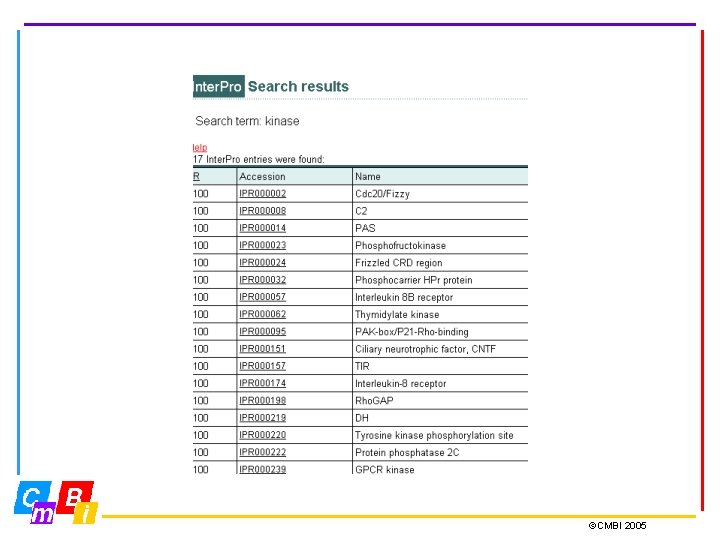
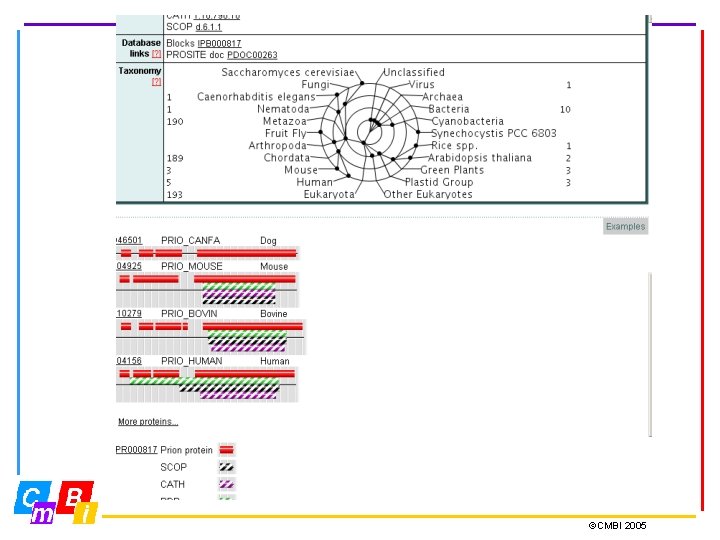
Integrating Pattern databases Inter. Pro - Integrated Documentation Resource of Protein Families, Domains and Functional Sites. Inter. Pro is a database of protein families, domains and functional sites in which identifiable features found in known proteins can be applied to unknown protein sequences. The aim is to provide a one-stop-shop for protein family diagnostics ©CMBI 2005

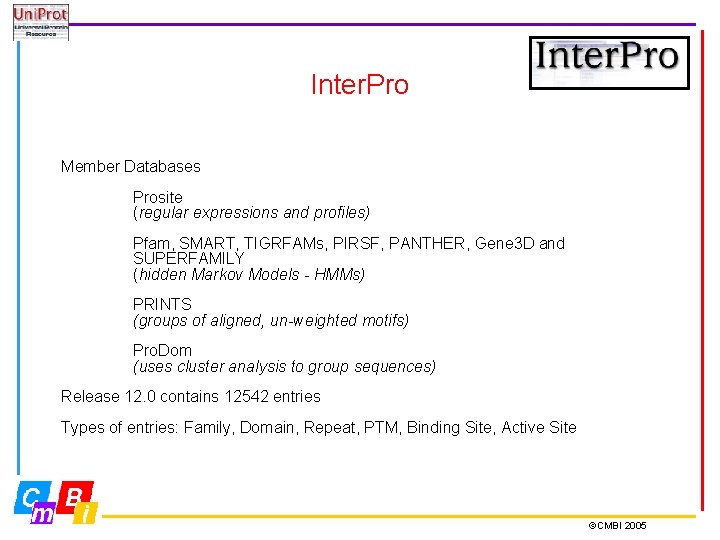
Inter. Pro Member Databases Prosite (regular expressions and profiles) Pfam, SMART, TIGRFAMs, PIRSF, PANTHER, Gene 3 D and SUPERFAMILY (hidden Markov Models - HMMs) PRINTS (groups of aligned, un-weighted motifs) Pro. Dom (uses cluster analysis to group sequences) Release 12. 0 contains 12542 entries Types of entries: Family, Domain, Repeat, PTM, Binding Site, Active Site ©CMBI 2005

©CMBI 2005

©CMBI 2005

©CMBI 2005

Summary • Many different protein signature databases exist (from small patterns to alignments to complex HMMs) • The databases have different strengths and weaknesses. Some databases can be better for your sequence than others • Therefore: best to combine methods, preferably in an integrated database • The quality of a database/server is best tested with a sequence you know very well • Always do control experiments: never trust a server ©CMBI 2005