Explorations of Multidimensional Sequence Space one symbol 1
Explorations of Multidimensional Sequence Space
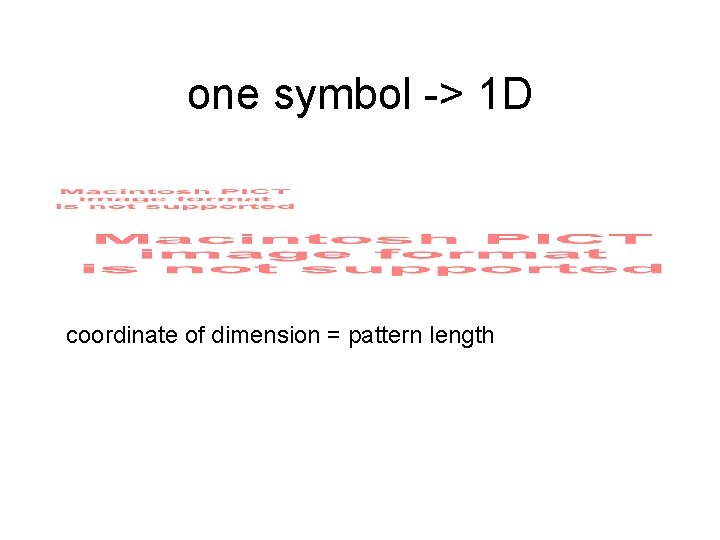
one symbol -> 1 D coordinate of dimension = pattern length
Two symbols -> Dimension = length of pattern length 1 = 1 D:
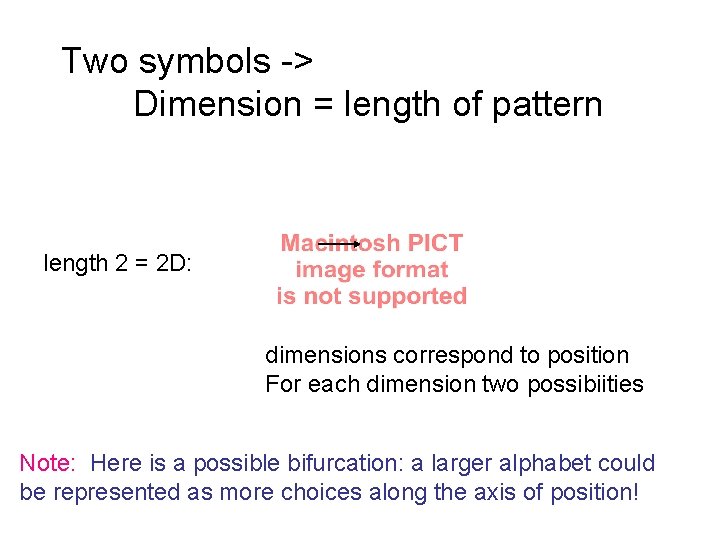
Two symbols -> Dimension = length of pattern length 2 = 2 D: dimensions correspond to position For each dimension two possibiities Note: Here is a possible bifurcation: a larger alphabet could be represented as more choices along the axis of position!
Two symbols -> Dimension = length of pattern length 3 = 3 D:
Two symbols -> Dimension = length of pattern length 4 = 4 D: aka Hypercube
Two symbols -> Dimension = length of pattern

Three Symbols (another solution is to use more values for each dimension)

Four Symbols: I. e. : with an alphabet of 4, we have a hypercube (4 D) already with a pattern size of 2, provided we stick to a binary pattern in each dimension.
hypercubes at 2 and 4 alphabets 2 character alphabet, pattern size 4 4 character alphabet, pattern size 2

Three Symbols Alphabet suggests fractal representation
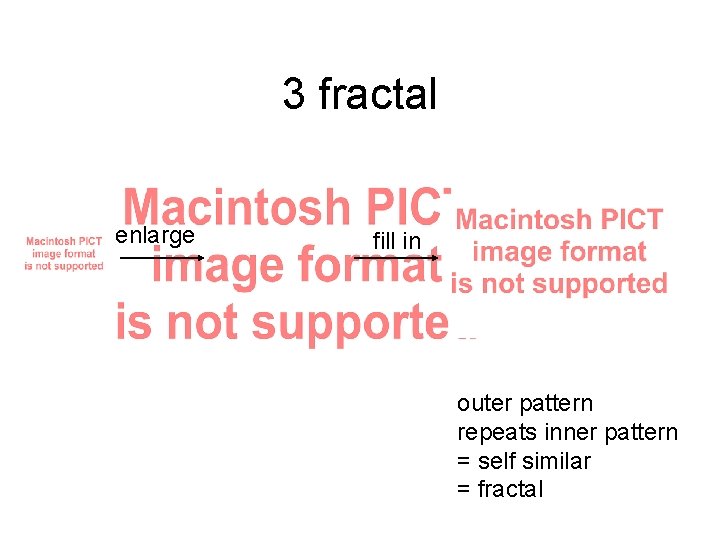
3 fractal enlarge fill in outer pattern repeats inner pattern = self similar = fractal

3 character alphapet 3 pattern fractal
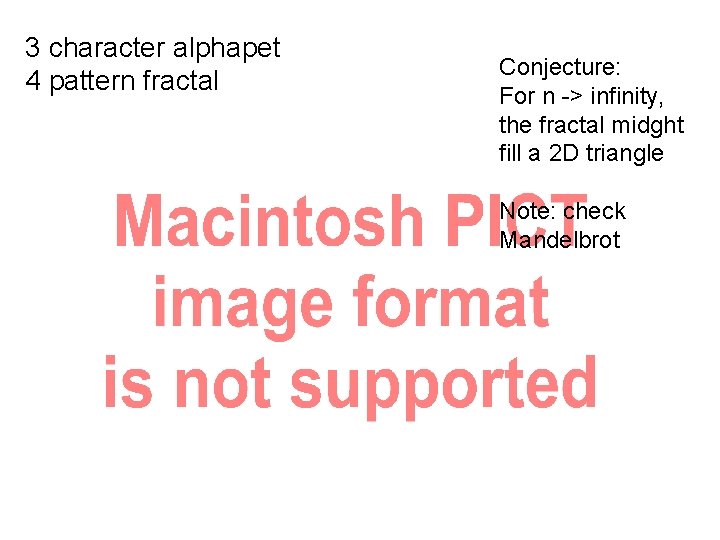
3 character alphapet 4 pattern fractal Conjecture: For n -> infinity, the fractal midght fill a 2 D triangle Note: check Mandelbrot
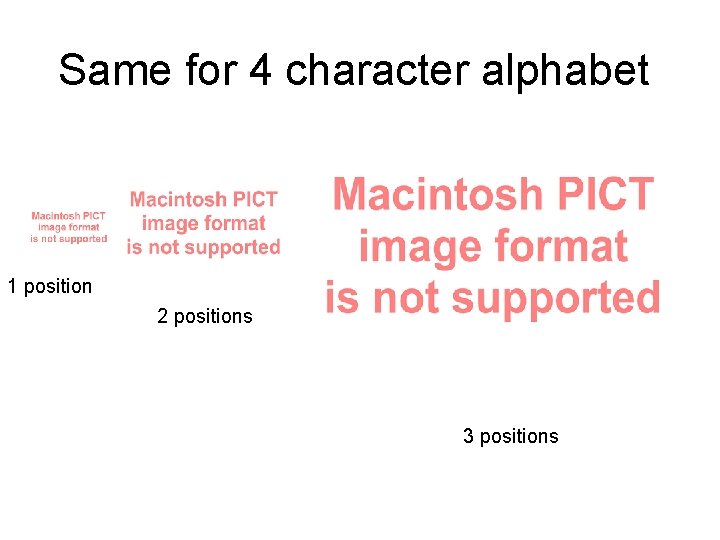
Same for 4 character alphabet 1 position 2 positions 3 positions

4 character alphabet continued (with cheating I didn’t actually add beads) 4 positions

4 character alphabet continued (with cheating I didn’t actually add beads) 5 positions

4 character alphabet continued (with cheating I didn’t actually add beads) 6 positions

4 character alphabet continued (with cheating I didn’t actually add beads) 7 positions
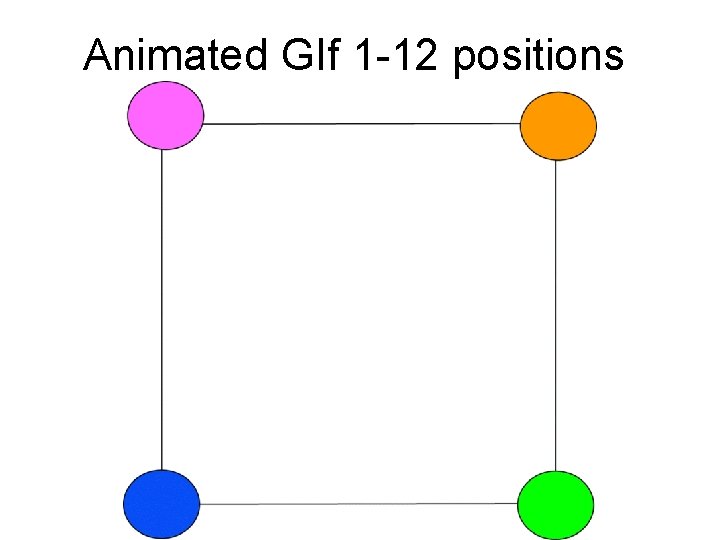
Animated GIf 1 -12 positions

Protein Space in Jal. View
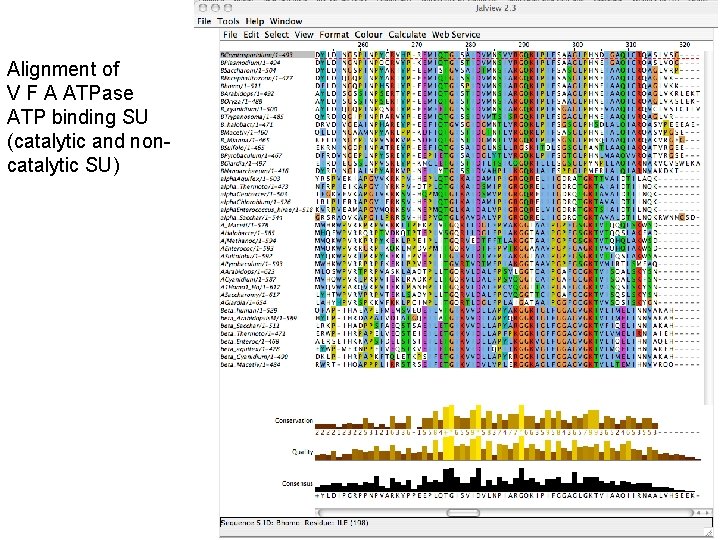
Alignment of V F A ATPase ATP binding SU (catalytic and noncatalytic SU)
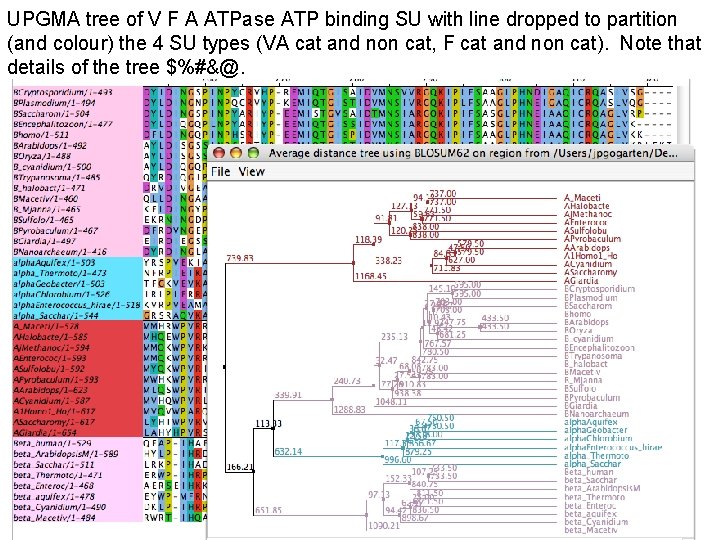
UPGMA tree of V F A ATPase ATP binding SU with line dropped to partition (and colour) the 4 SU types (VA cat and non cat, F cat and non cat). Note that details of the tree $%#&@.
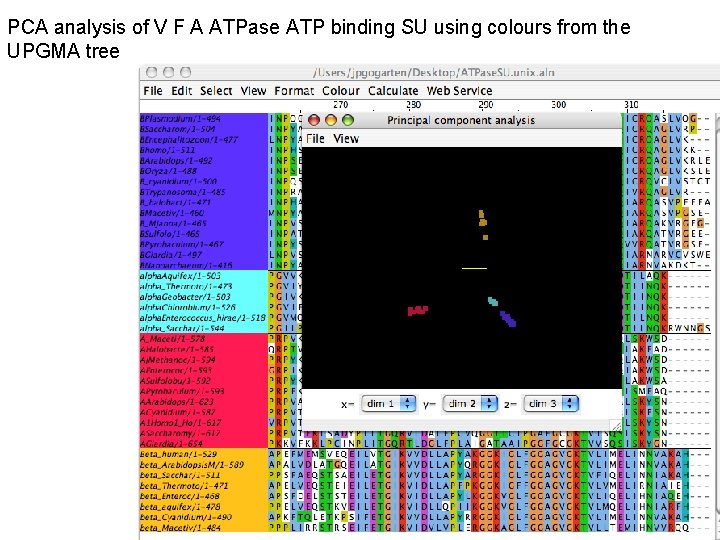
PCA analysis of V F A ATPase ATP binding SU using colours from the UPGMA tree
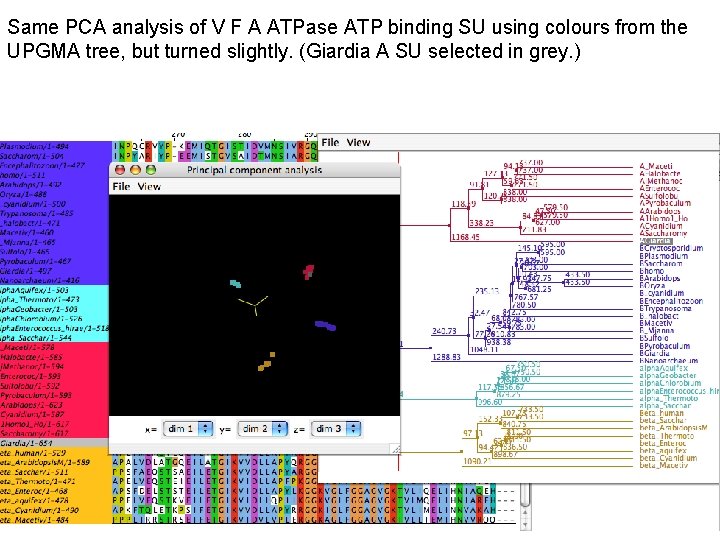
Same PCA analysis of V F A ATPase ATP binding SU using colours from the UPGMA tree, but turned slightly. (Giardia A SU selected in grey. )
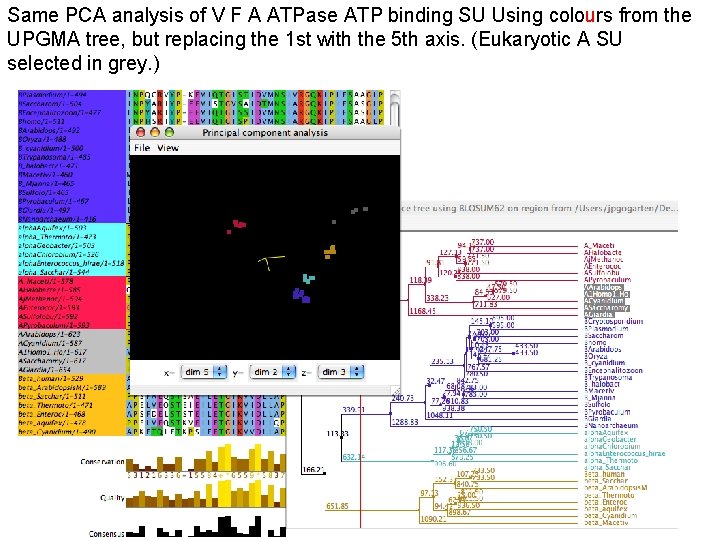
Same PCA analysis of V F A ATPase ATP binding SU Using colours from the UPGMA tree, but replacing the 1 st with the 5 th axis. (Eukaryotic A SU selected in grey. )
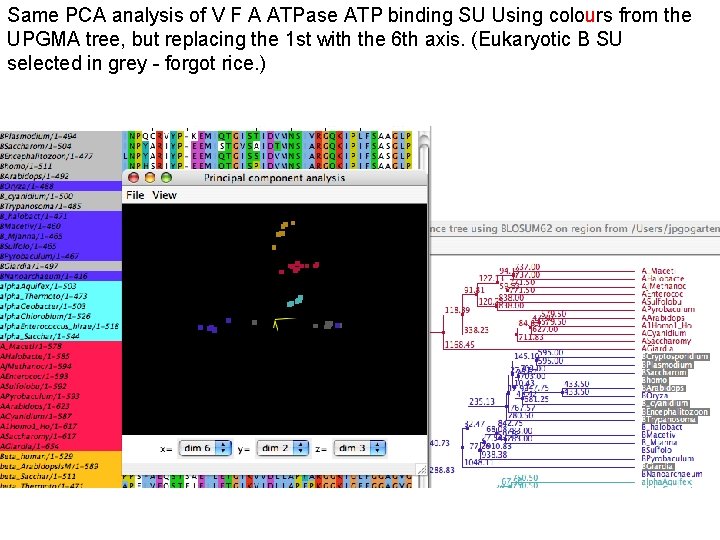
Same PCA analysis of V F A ATPase ATP binding SU Using colours from the UPGMA tree, but replacing the 1 st with the 6 th axis. (Eukaryotic B SU selected in grey - forgot rice. )

Problems • Jalview’s approach requires an alignment. • Solution: Use pattern absence / presence as coordinate • Which patterns? – GBLOCKS (new additions use PSSMs) – CDD PSSM profiles – It would be nice to stick to small words. • One could screen for words/motifs/PSSMs that have a good power of resolution: – PCA with all, choose only the ones that contribute to the main axis – probably better to do data bank search and find how often it is present. One could generate random motifs (or all possible motifs) and check them out (Criterion needs work). – Empirical orthogonality – Exhaustive vs random – How to judge discriminatory power (maybe 5% significance value) – Present absence - optimal discriminatory power?
- Slides: 28