Expanded Representation of EC Numbers in Pathway Tools

Expanded Representation of EC Numbers in Pathway Tools Suzanne Paley March 2013 1 SRI International Bioinformatics

Motivations EC: Enzyme Commission system for identifying enzyme functionality l Multifunctional enzyme should have different EC# for each function l Old Pathway Tools representation: l Assumption: single function of an enzyme = single reaction l Each reaction was assigned a single EC# l An enzyme has EC#s derived from all reactions it catalyzes l Problems: l Enzyme Commission can assign multiple EC#s to same reaction due to different cellular roles, different substrate specificities, different cofactors, etc. l A single EC# can refer to multiple reactions due to substrate non-specificity, sub-reactions, etc. l 1: 1 mapping between EC#s and reactions was increasingly causing problems l 2 SRI International Bioinformatics

New Representation (v. 16. 5) Reaction can have multiple EC#s l New EC-Numbers database l Object created for every EC# (and every EC class) l Contains name, synonyms, comments, citations from Enzyme Commission l Maintained by. Meta. Cyc curators, loaded by every PGDB l Reaction object in a PGDB points to one or more EC# objects in EC-Numbers database l Link to EC# can be annotated as an “official” EC reaction l u u Meta. Cyc contains every EC reaction, other PGDBs contain a subset SRI International Bioinformatics change l Non-backwards-compatible l 3 Official means it is part of the EC# definition from the Enzyme Commission There can be additional “unofficial” reactions associated with an EC#, e. g. for alternate substrates, specific substrates, etc.
Associating EC# With Enzyme l Enzymes not directly linked to EC#s – EC#s are inferred from reactions they catalyze l An enzyme must catalyze all “official” EC reactions for a given EC# in order to be associated w/ that EC# l May or may not catalyze “unofficial” EC reactions l If an enzyme catalyzes multiple reactions, it is preferentially associated with EC# that covers all the reactions, rather than EC#s of individual reactions 4 SRI International Bioinformatics

Example: 1. 1. 6/1. 1. 21 l 1. 1. 6 l -- catalase 2 H 2 O 2 -> 2 H 2 O + O 2 l 1. 1. 21 l l – catalase-peroxidase 2 H 2 O 2 -> 2 H 2 O + O 2 2 nd reaction uses generic electron acceptor l Catalase reaction assigned both EC#s l kat. E only catalyzes first reaction, inferred EC 1. 1. 6 l kat. G catalyzes both reactions, inferred EC 1. 1. 21, but not 1. 1. 6 5 SRI International Bioinformatics
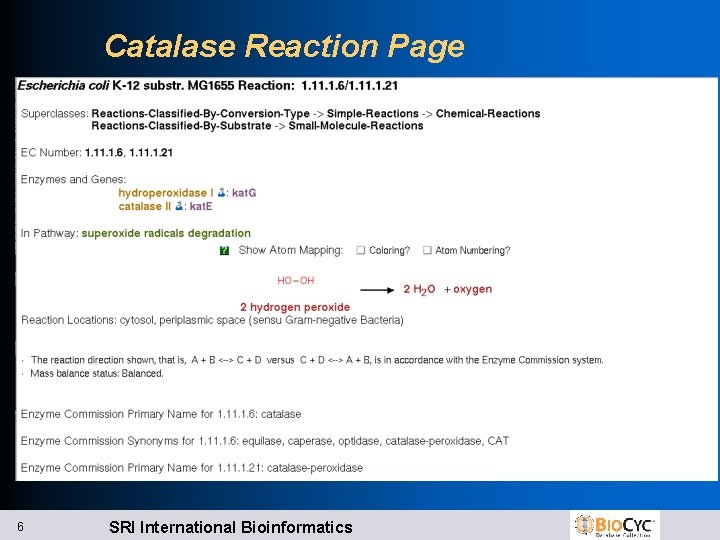
Catalase Reaction Page 6 SRI International Bioinformatics

EC Number Pages 7 SRI International Bioinformatics
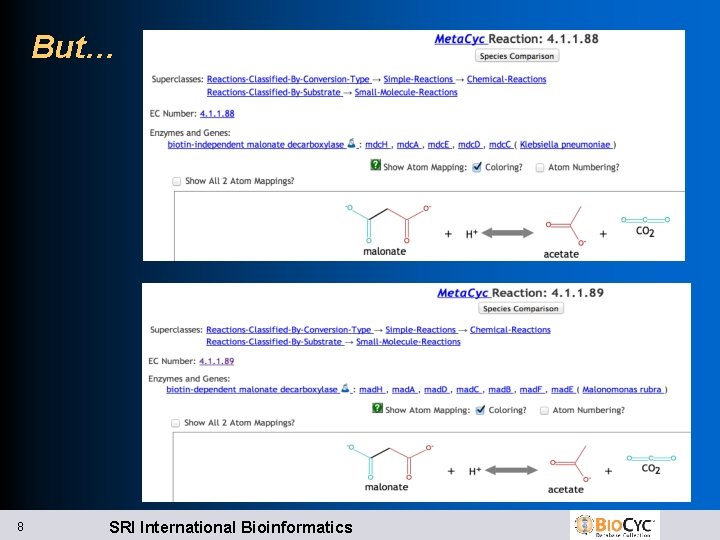
But… 8 SRI International Bioinformatics
Other Changes: Enzyme Kinetic Data l Kinetic parameters Km, kcat, Vmax, specific activity associated with enzymatic-reaction l Km also associated with particular substrate in enzymatic-reaction l Recent change: Now all parameters are also associated with substrate in enzymatic-reaction l Enables us to compute kcat/Km (catalytic activity) l Developed program to infer appropriate substrate for 400+ kinetic data values previously entered w/o substrate in Eco. Cyc and Meta. Cyc 9 SRI International Bioinformatics
- Slides: 9