Example Calculation of genetic distance Problem Calculation of
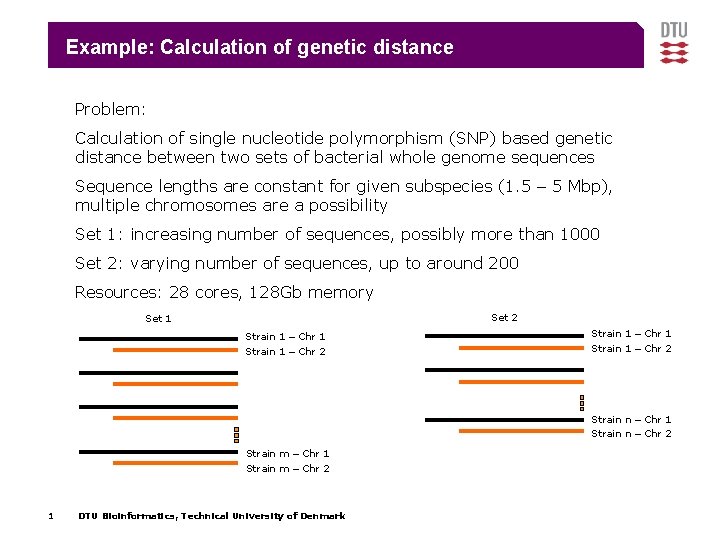
Example: Calculation of genetic distance Problem: Calculation of single nucleotide polymorphism (SNP) based genetic distance between two sets of bacterial whole genome sequences Sequence lengths are constant for given subspecies (1. 5 – 5 Mbp), multiple chromosomes are a possibility Set 1: increasing number of sequences, possibly more than 1000 Set 2: varying number of sequences, up to around 200 Resources: 28 cores, 128 Gb memory Set 2 Set 1 Strain 1 – Chr 2 Strain 1 – Chr 1 Strain 1 – Chr 2 Strain n – Chr 1 Strain n – Chr 2 Strain m – Chr 1 Strain m – Chr 2 1 DTU Bioinformatics, Technical University of Denmark
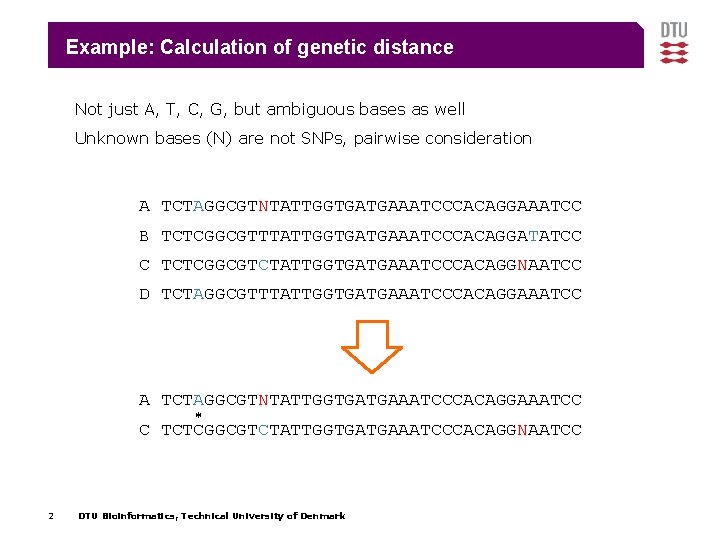
Example: Calculation of genetic distance Not just A, T, C, G, but ambiguous bases as well Unknown bases (N) are not SNPs, pairwise consideration A TCTAGGCGTNTATTGGTGATGAAATCCCACAGGAAATCC B TCTCGGCGTTTATTGGTGATGAAATCCCACAGGATATCC C TCTCGGCGTCTATTGGTGATGAAATCCCACAGGNAATCC D TCTAGGCGTTTATTGGTGATGAAATCCCACAGGAAATCC A TCTAGGCGTNTATTGGTGATGAAATCCCACAGGAAATCC * C TCTCGGCGTCTATTGGTGATGAAATCCCACAGGNAATCC 2 DTU Bioinformatics, Technical University of Denmark
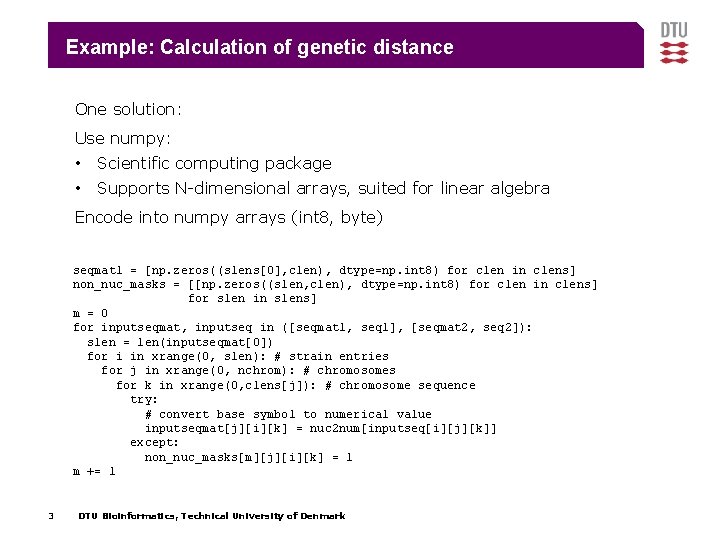
Example: Calculation of genetic distance One solution: Use numpy: • Scientific computing package • Supports N-dimensional arrays, suited for linear algebra Encode into numpy arrays (int 8, byte) seqmat 1 = [np. zeros((slens[0], clen), dtype=np. int 8) for clen in clens] non_nuc_masks = [[np. zeros((slen, clen), dtype=np. int 8) for clen in clens] for slen in slens] m = 0 for inputseqmat, inputseq in ([seqmat 1, seq 1], [seqmat 2, seq 2]): slen = len(inputseqmat[0]) for i in xrange(0, slen): # strain entries for j in xrange(0, nchrom): # chromosomes for k in xrange(0, clens[j]): # chromosome sequence try: # convert base symbol to numerical value inputseqmat[j][i][k] = nuc 2 num[inputseq[i][j][k]] except: non_nuc_masks[m][j][i][k] = 1 m += 1 3 DTU Bioinformatics, Technical University of Denmark
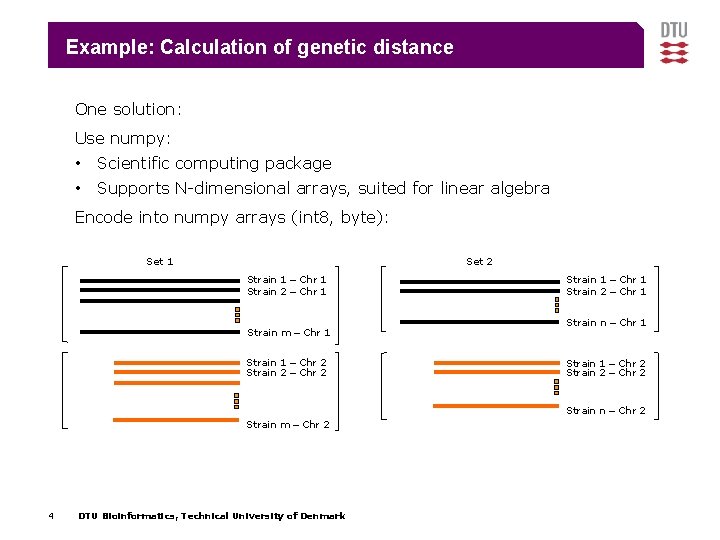
Example: Calculation of genetic distance One solution: Use numpy: • Scientific computing package • Supports N-dimensional arrays, suited for linear algebra Encode into numpy arrays (int 8, byte): Set 1 Set 2 Strain 1 – Chr 1 Strain 2 – Chr 1 Strain m – Chr 1 Strain 1 – Chr 2 Strain 2 – Chr 2 Strain 1 – Chr 1 Strain 2 – Chr 1 Strain n – Chr 1 Strain 1 – Chr 2 Strain 2 – Chr 2 Strain n – Chr 2 Strain m – Chr 2 4 DTU Bioinformatics, Technical University of Denmark
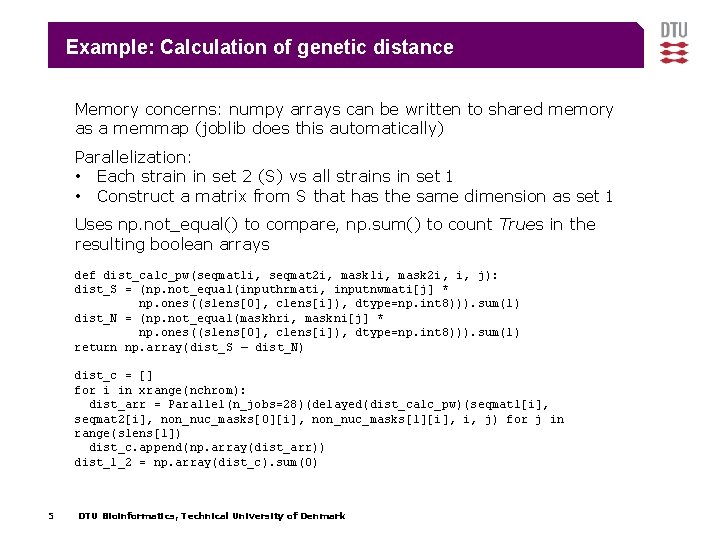
Example: Calculation of genetic distance Memory concerns: numpy arrays can be written to shared memory as a memmap (joblib does this automatically) Parallelization: • Each strain in set 2 (S) vs all strains in set 1 • Construct a matrix from S that has the same dimension as set 1 Uses np. not_equal() to compare, np. sum() to count Trues in the resulting boolean arrays def dist_calc_pw(seqmat 1 i, seqmat 2 i, mask 1 i, mask 2 i, i, j): dist_S = (np. not_equal(inputhrmati, inputnwmati[j] * np. ones((slens[0], clens[i]), dtype=np. int 8))). sum(1) dist_N = (np. not_equal(maskhri, maskni[j] * np. ones((slens[0], clens[i]), dtype=np. int 8))). sum(1) return np. array(dist_S – dist_N) dist_c = [] for i in xrange(nchrom): dist_arr = Parallel(n_jobs=28)(delayed(dist_calc_pw)(seqmat 1[i], seqmat 2[i], non_nuc_masks[0][i], non_nuc_masks[1][i], i, j) for j in range(slens[1]) dist_c. append(np. array(dist_arr)) dist_1_2 = np. array(dist_c). sum(0) 5 DTU Bioinformatics, Technical University of Denmark
- Slides: 5