Evolutionary Trace Intuition and Applications COMBINING THE EVOLUTIONARY
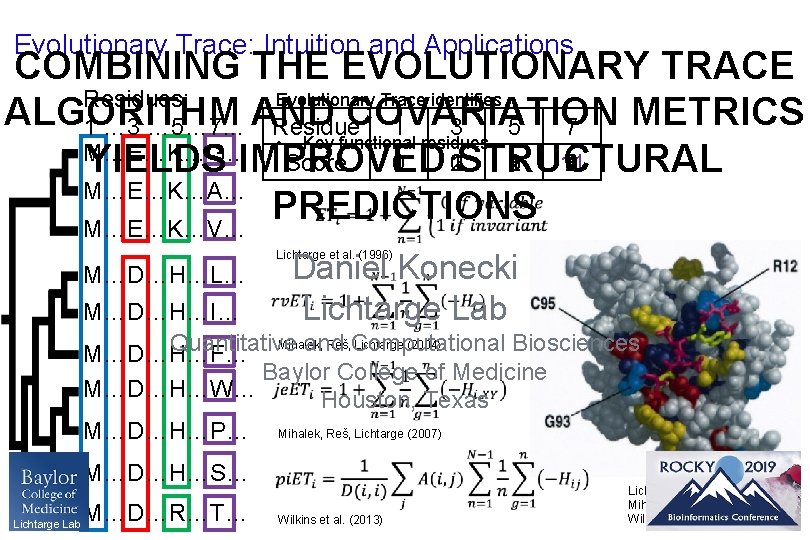
Evolutionary Trace: Intuition and Applications COMBINING THE EVOLUTIONARY TRACE Evolutionary Trace identifies Residues: ALGORITHM COVARIATION METRICS Residue 1 3 5 7 1 … 3 … 5… 7… AND • Key functional residues M…E…K…G… Score 0 0 1 2 STRUCTURAL 1 3 2 1 11 4 3 2 1 YIELDS IMPROVED M…E…K…A… • Binding interfaces PREDICTIONS M…E…K…V… • Active sites Lichtarge et al. (1996) M…D…H…L… Daniel Konecki Lichtarge Lab M…D…H…I… Mihalek, Reš, Lichtarge (2004) Quantitative and Computational Biosciences M…D…H…F… Baylor College of Medicine M…D…H…W… Houston, Texas M…D…H…P… Mihalek, Reš, Lichtarge (2007) M…D…H…S… Lichtarge Lab M…D…R…T… Wilkins et al. (2013) Lichtarge et al. (1996) Mihalek, Reš, Lichtarge (2004) Wilkins et al. (2013)
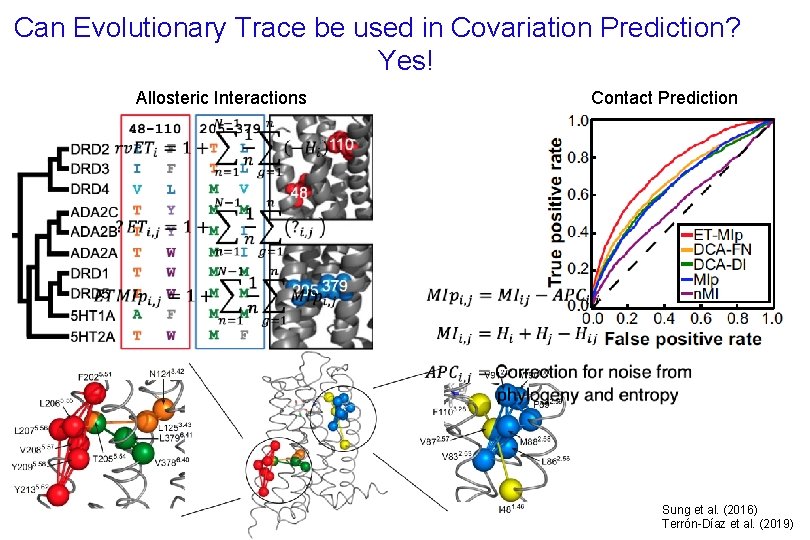
Can Evolutionary Trace be used in Covariation Prediction? Yes! Contact Prediction Allosteric Interactions Sung et al. (2016) Terrón-Díaz et al. (2019)
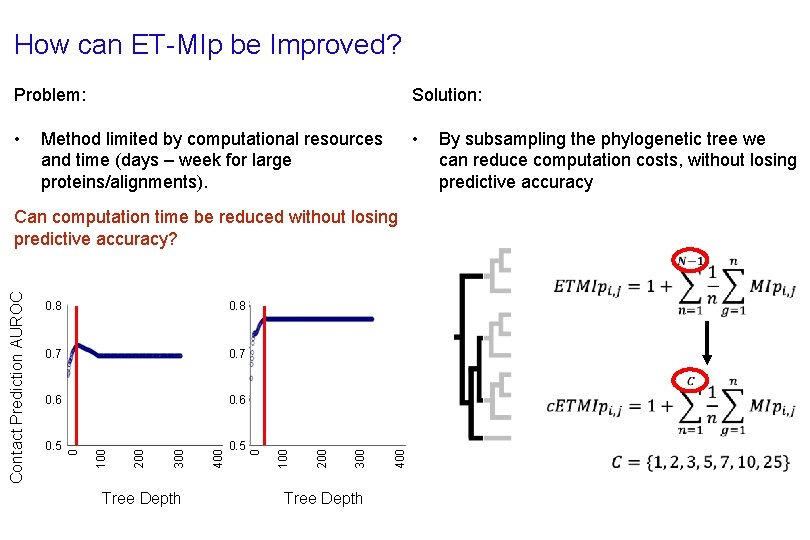
How can ET-MIp be Improved? Problem: Solution: • • Method limited by computational resources and time (days – week for large proteins/alignments). By subsampling the phylogenetic tree we can reduce computation costs, without losing predictive accuracy Can computation time be reduced without losing predictive accuracy? 0. 8 0. 7 0. 6 0. 5 Tree Depth 400 300 200 100 0 Contact Prediction AUROC
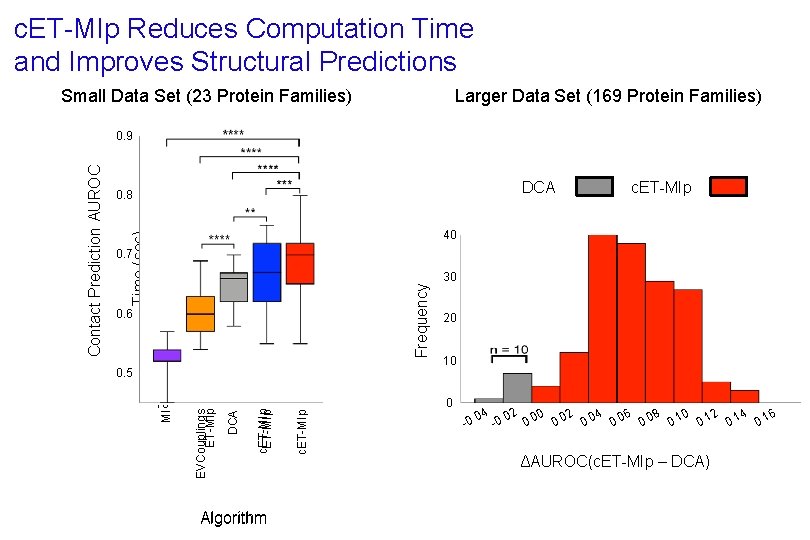
c. ET-MIp Reduces Computation Time and Improves Structural Predictions Small Data Set (23 Protein Families) 106 DCA 0. 8 Time (sec) 104 c. ET-MIp 40 0. 6 Frequency 0. 7 102 Algorithm 30 20 10 0 c. ET-MIp DCA 100 EVCouplings ET-MIp 0. 5 MI Contact Prediction AUROC 0. 9 Larger Data Set (169 Protein Families) 04 0. 02 0. 00 0. 02 0. 04 0. 06 0. 08 0. 10 0. 12 0. 14 0. 16 - -0. ΔAUROC(c. ET-MIp – DCA)
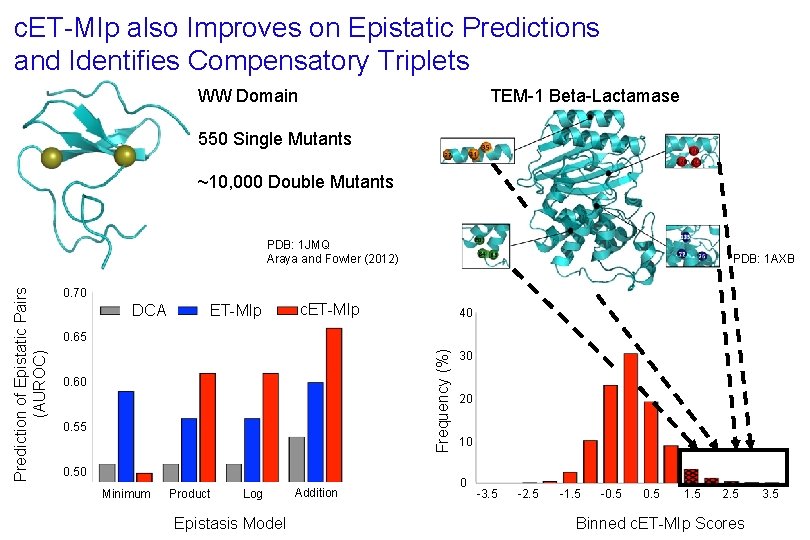
c. ET-MIp also Improves on Epistatic Predictions and Identifies Compensatory Triplets TEM-1 Beta-Lactamase WW Domain 550 Single Mutants ~10, 000 Double Mutants PDB: 1 AXB 0. 70 DCA ET-MIp c. ET-MIp 40 0. 65 Frequency (%) Prediction of Epistatic Pairs (AUROC) PDB: 1 JMQ Araya and Fowler (2012) 0. 60 0. 55 0. 50 Minimum Product Log Epistasis Model Addition 30 20 10 0 -3. 5 -2. 5 -1. 5 -0. 5 1. 5 2. 5 Binned c. ET-MIp Scores 3. 5

Conclusions and Implications Conclusions: Implications: • Evolutionary Trace improves covariation predictions • Better covariation predictions may be possible if Evolutionary Trace is combined with other covariation metrics • Most improvement is observed early in the trace • These predictions provide orthogonal features for ML pipelines used for ab initio structural modeling • These predictions provide a rich feature with the potential to improve ML models predicting protein structure • Structure and function prediction are improved by sub-sampling the tree Thank You! Please visit poster #40 to discuss further! konecki@bcm. edu
- Slides: 6