Evolutionary Morphing and Shape Distance Nina Amenta Computer













- Slides: 36

Evolutionary Morphing and Shape Distance Nina Amenta Computer Science, UC Davis

Collaborators Physical Anthropology Eric Delson, Steve Frost, Lissa Tallman, Will Harcourt-Smith Morphometrics F. James Rohlf Computer Science and Math Katherine St. John, David Wiley, Deboshmita Ghosh, Misha Kazhdan, Owen Carmichael, Joel Hass, David Coeurjolly

Outline • Application of 3 D Procrustes tangent space analysis in primate evolution • Some issues with the shape space • An idea

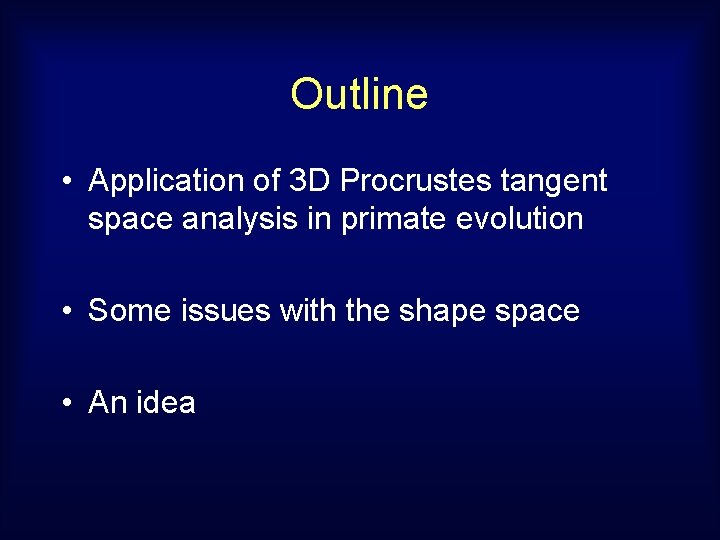
Evolutionary Trees

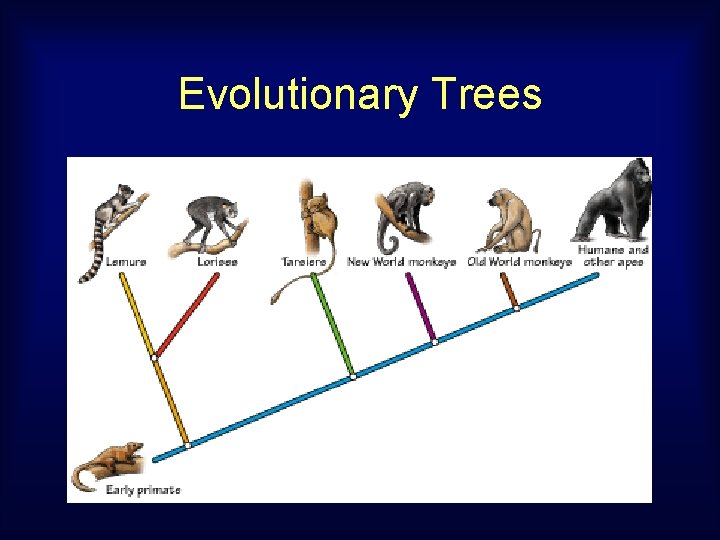
Computing Trees on extant species come from genomic data. Papio Macaca Cercocebus Cercopithecus Allenopithecus Tree inference method

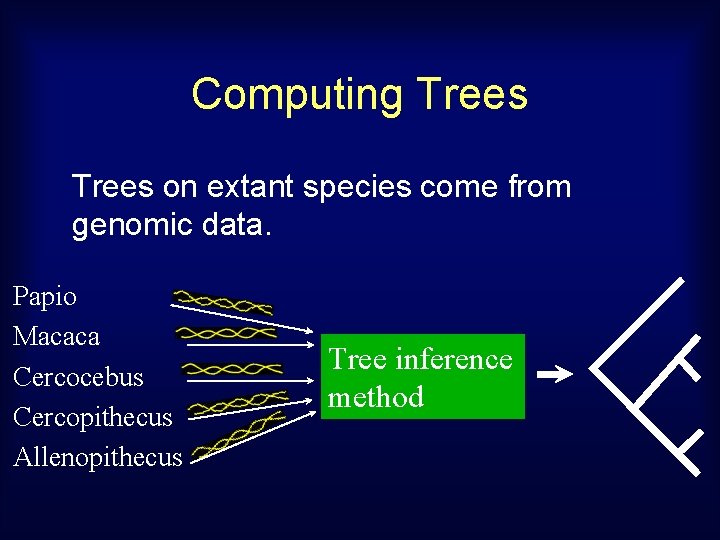
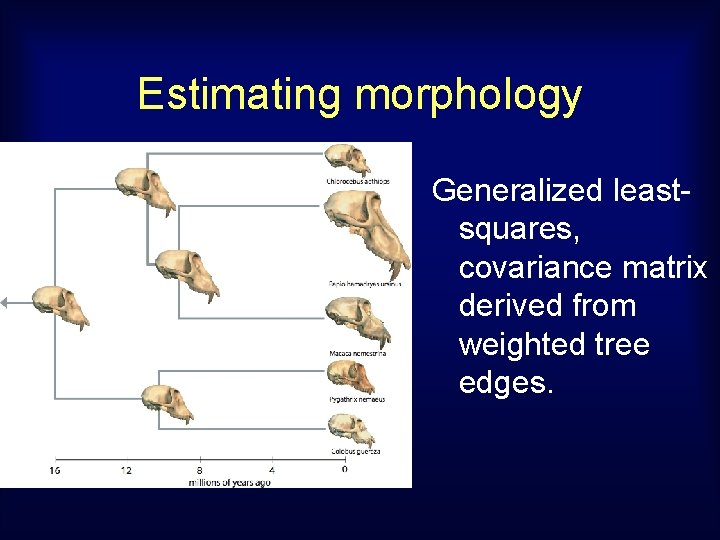
Estimating morphology Using 3 D data for extant species, and tree, estimate cranial shapes for the hypothetical ancestors. 3 D input data

Estimating morphology Generalized leastsquares, covariance matrix derived from weighted tree edges.

Evolutionary Morphing

Fossils Genomic trees don’t include fossils. Primates: ~200 extinct genera, ~60 extant. Fossils have to be added based on shape and meta-data.

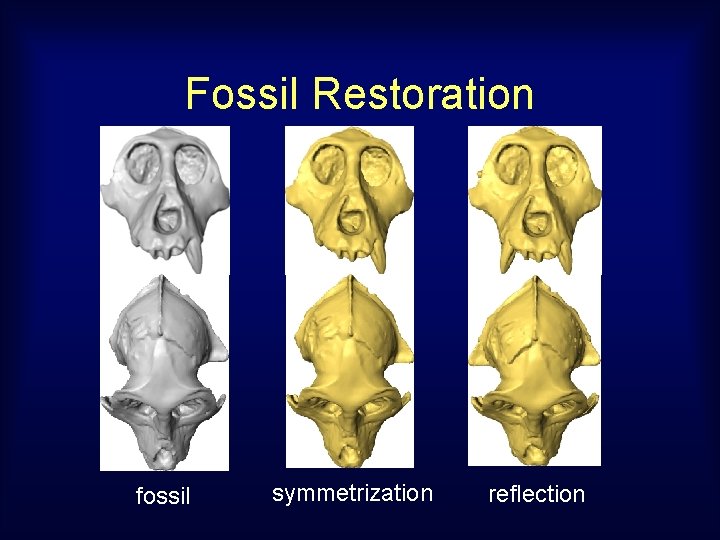
Fossil Restoration fossil symmetrization reflection

Sahelanthropos

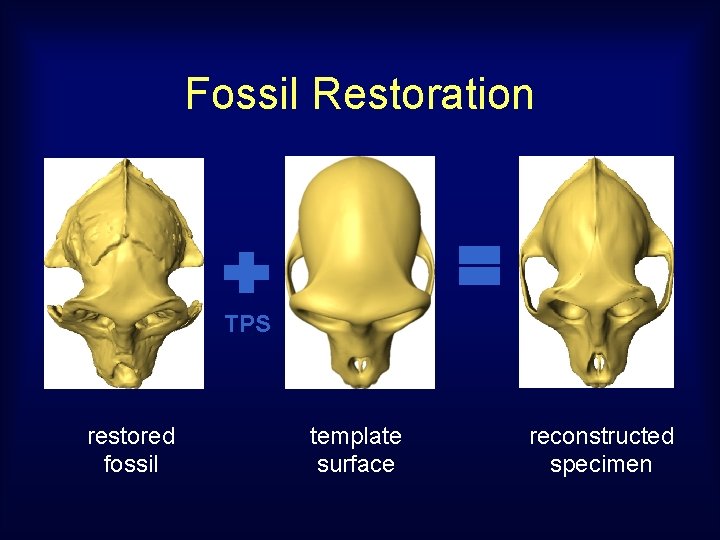
Fossil Restoration TPS restored fossil template surface reconstructed specimen

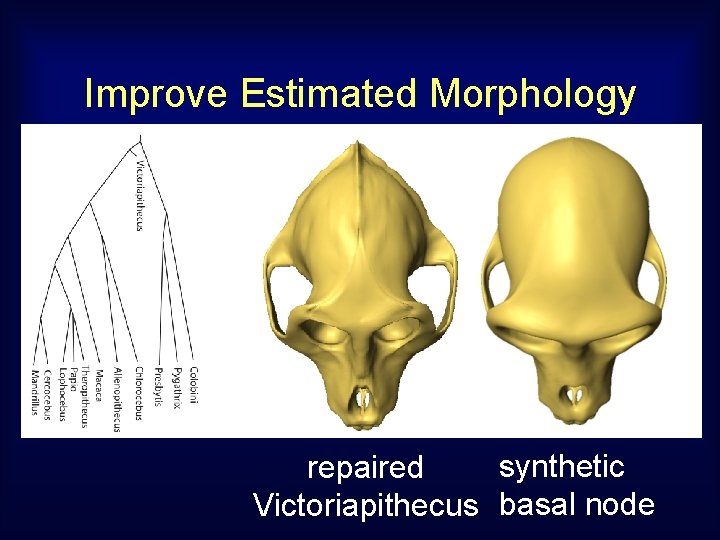
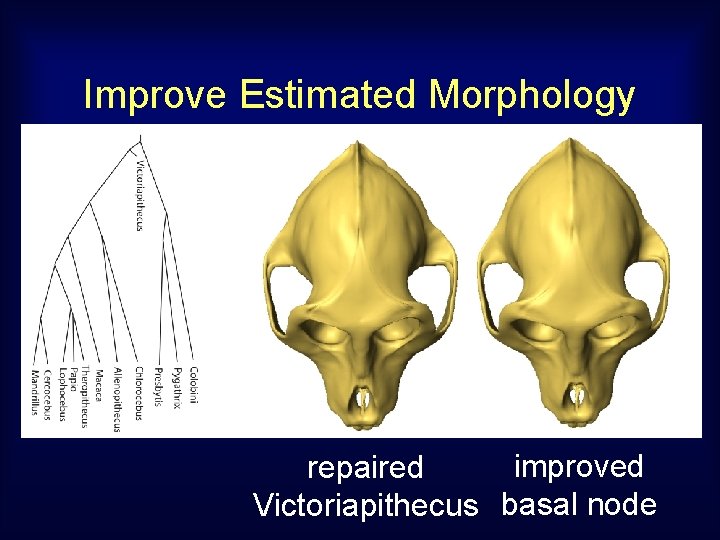
Improve Estimated Morphology synthetic repaired Victoriapithecus basal node

Improve Estimated Morphology improved repaired Victoriapithecus basal node

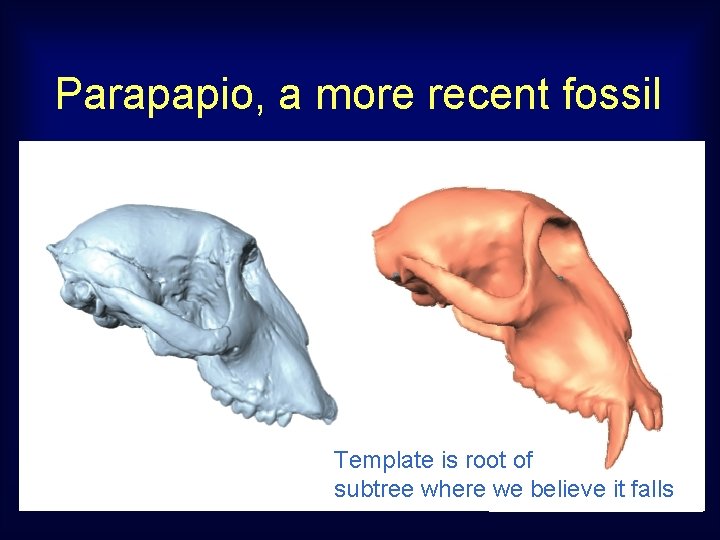
Parapapio, a more recent fossil Template is root of subtree where we believe it falls

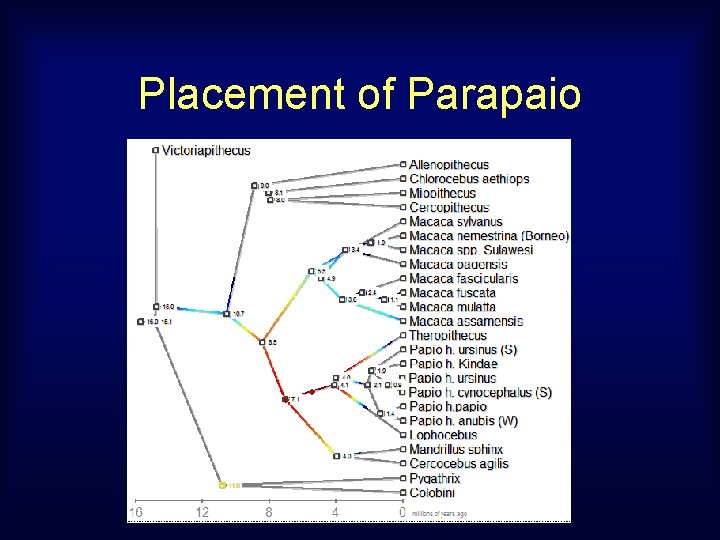
Placement of Parapaio

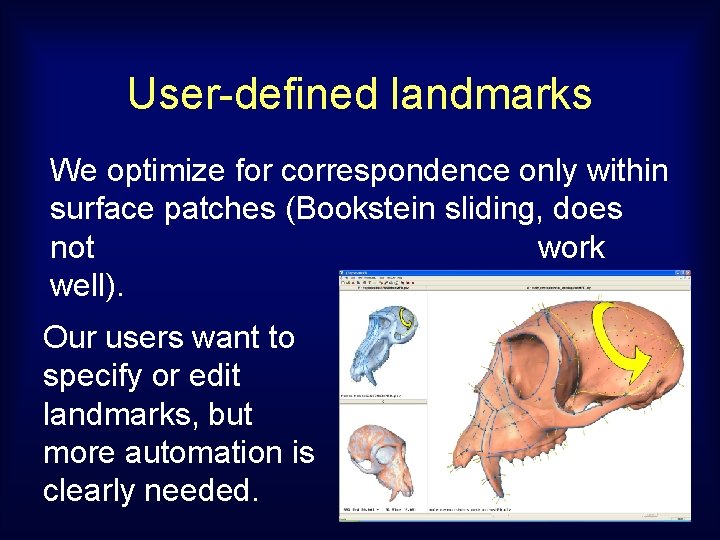
User-defined landmarks We optimize for correspondence only within surface patches (Bookstein sliding, does not work well). Our users want to specify or edit landmarks, but more automation is clearly needed.

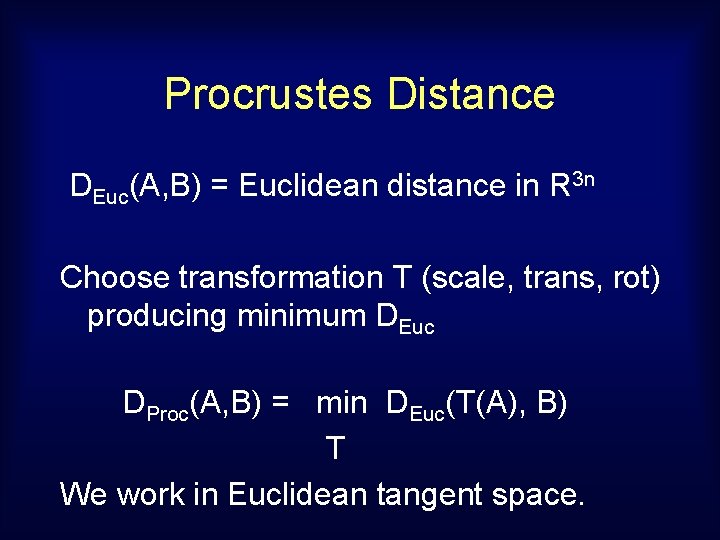
Procrustes Distance DEuc(A, B) = Euclidean distance in R 3 n Choose transformation T (scale, trans, rot) producing minimum DEuc DProc(A, B) = min DEuc(T(A), B) T We work in Euclidean tangent space.

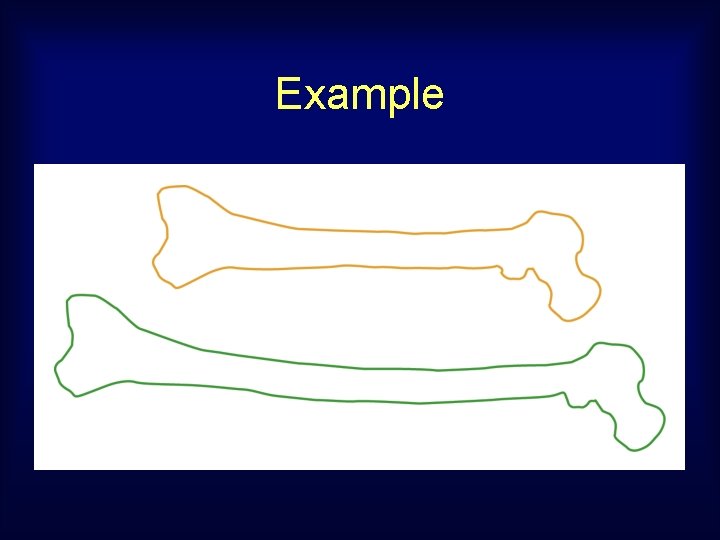
Example

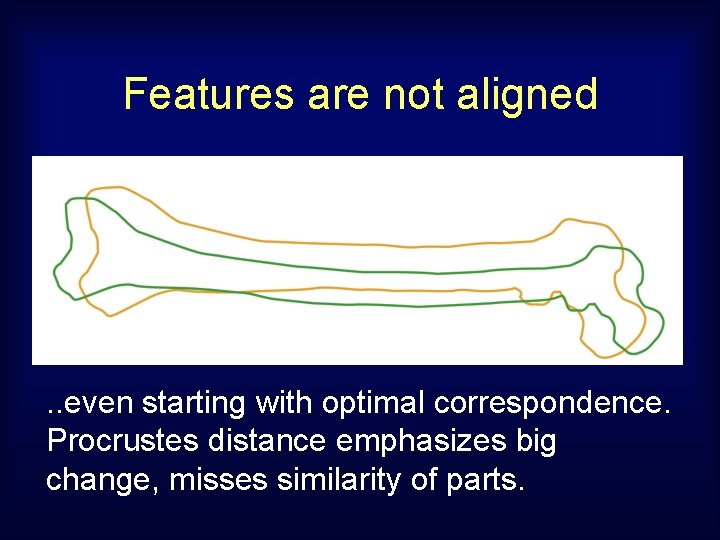
Features are not aligned . . even starting with optimal correspondence. Procrustes distance emphasizes big change, misses similarity of parts.

Features are not aligned Changing the details might even reduce DProc.

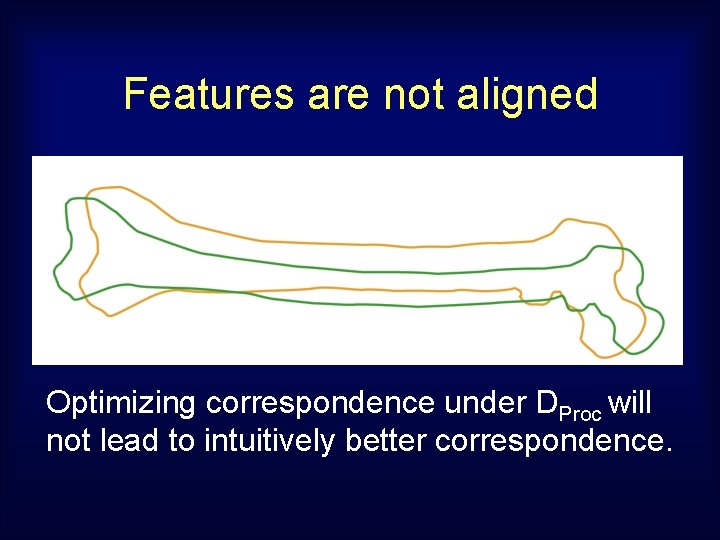
Features are not aligned Optimizing correspondence under DProc will not lead to intuitively better correspondence.

Complex Shapes All parts cannot be simultaneously aligned by linear deformations. Deformation really is non-linear.

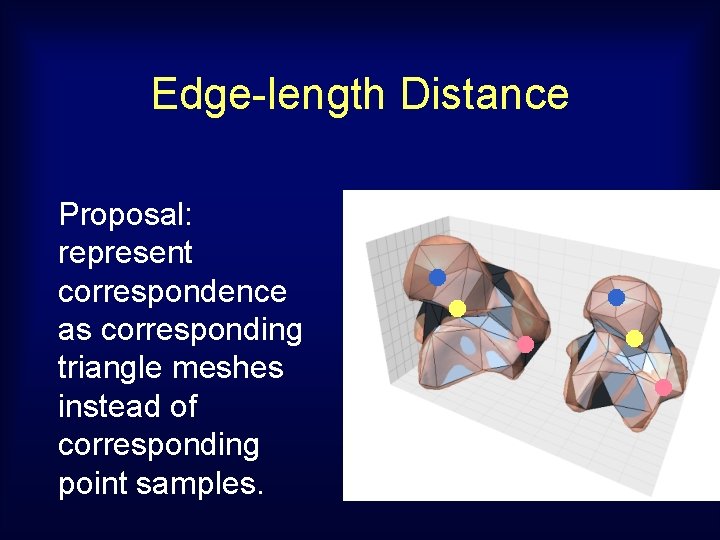
Edge-length Distance Proposal: represent correspondence as corresponding triangle meshes instead of corresponding point samples.

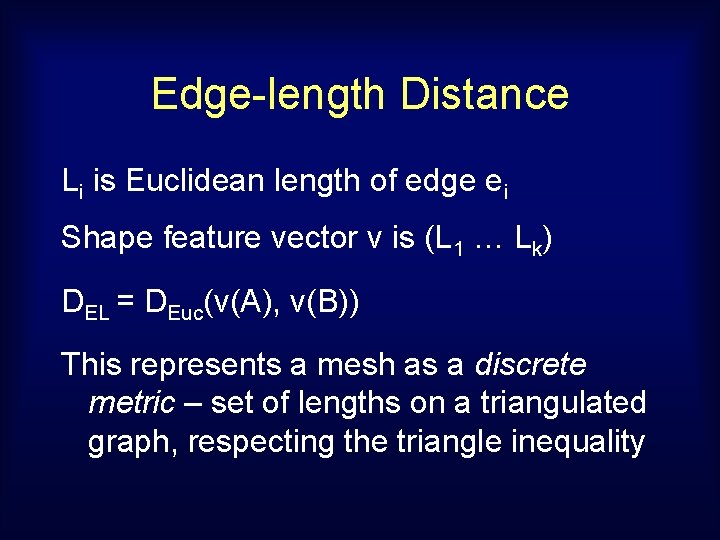
Edge-length Distance Li is Euclidean length of edge ei Shape feature vector v is (L 1 … Lk) DEL = DEuc(v(A), v(B)) This represents a mesh as a discrete metric – set of lengths on a triangulated graph, respecting the triangle inequality

Information Loss In 2 D, this does not make much sense. But in 3 D, almost all triangulated polyhedra are rigid. So a discrete metric has a finite number of rigid realizations.

Not a New Idea Euclidean Distance Matrix Analysis, Lele and Richtmeier, 2001 – use the complete distance matrix as shape rep. “Truss metrics” – include only enough edges to get rigidity.

Quote “…the arbitrary choice of a subset of linear distances could accentuate the influence of certain linear distances in the comparison of forms, while masking the influence of others. ” - Richtsmeier, Deleon, and Lele, 2002. Not an issue in R 3!

Nice Properties • Rotation and translation invariant • Invariant to rotations and translations of parts (isometries). • Any convex combination of specimens gives another vector of Li obeying triangle inequalities. So we can do statistics in a convex region of Euclidean space.

Scale Can normalize to produce scale invariance, as with Procrustes distance. Choosing scale so that S Li = 1 keeps all specimens in a linear subspace.

Degrees of Freedom Dimension of Kendall shape space is 3 n-7 Number of edges for a triangulated object homeomorphic to a sphere is 3 n-6 (Euler+triangulation constraints), -1 for scale = 3 n-7

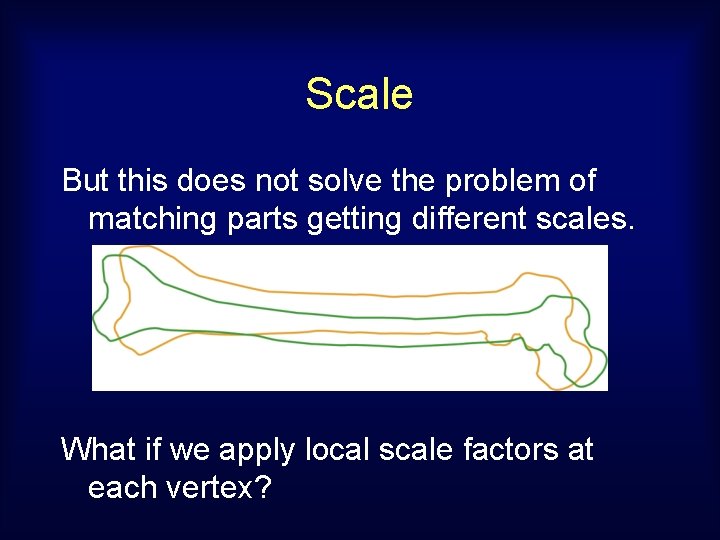
Scale But this does not solve the problem of matching parts getting different scales. What if we apply local scale factors at each vertex?

Local Scale? We could add a scale factor at each vertex, producing a discrete conformal representation (Springborn, Schoeder, Bobenko, Pinkall)…but this has way too many degrees of freedom. Q 1: How to incorporate the right amount of local scale?

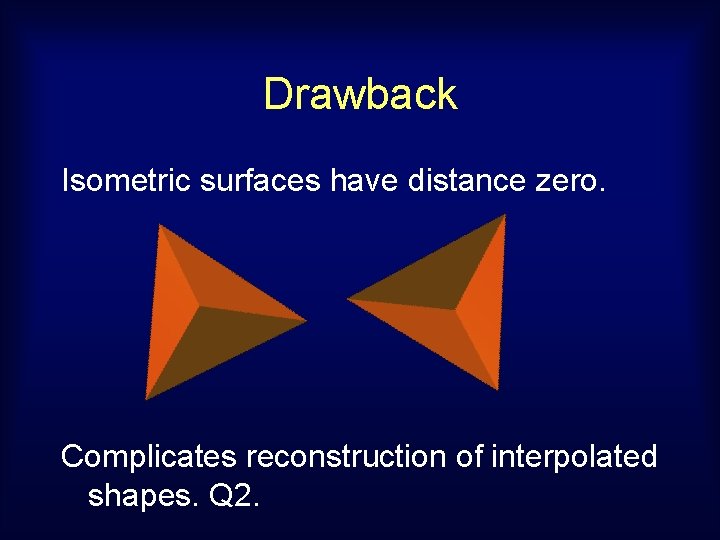
Drawback Isometric surfaces have distance zero. Complicates reconstruction of interpolated shapes. Q 2.

More Questions Q 3: Given a discrete metric formed as a convex combination of specimens, how to choose the right 3 D realization for visualization? Q 4: How to optimize correspondence so as to minimize DEL? How to weight by area?

Thank you!