Evolution Time Travel Genome Evolution Evolution Time Travel



![Definitions • [F. Farnoud, M. Schwartz, J. Bruck, ISIT’ 14] Definitions • [F. Farnoud, M. Schwartz, J. Bruck, ISIT’ 14]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-23.jpg)
![[S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017] [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-34.jpg)
![Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017] Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-35.jpg)
![Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017] Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-36.jpg)




![[S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839] [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-92.jpg)
![[S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839] [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-93.jpg)
![Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Class_4 = [rest] [S. Jain, Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Class_4 = [rest] [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-94.jpg)
![Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-95.jpg)
![Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-96.jpg)
![Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-97.jpg)





- Slides: 104


Evolution Time Travel

Genome Evolution ? Evolution Time Travel Me

Decoding the Past Lecture 17

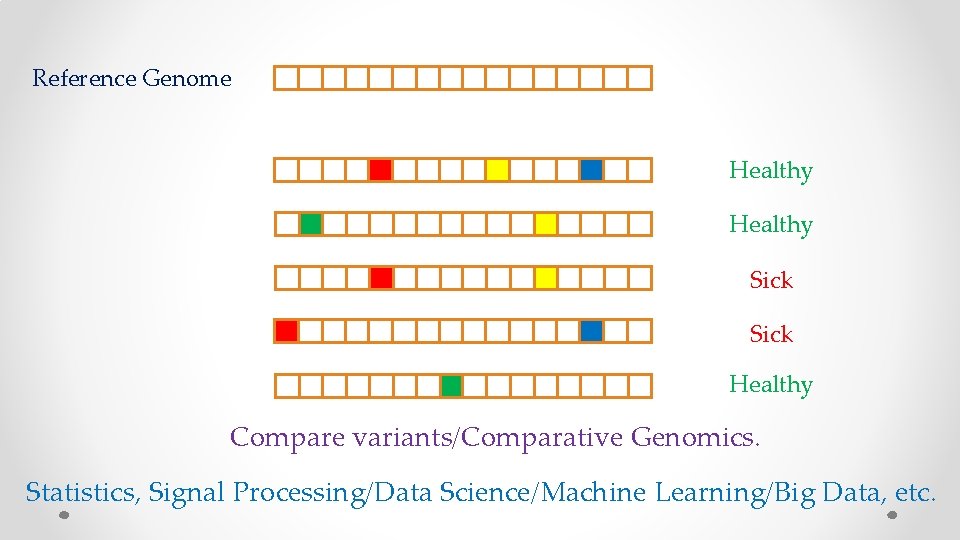
Reference Genome

Reference Genome Healthy Sick Healthy Compare variants/Comparative Genomics. Statistics, Signal Processing/Data Science/Machine Learning/Big Data, etc.

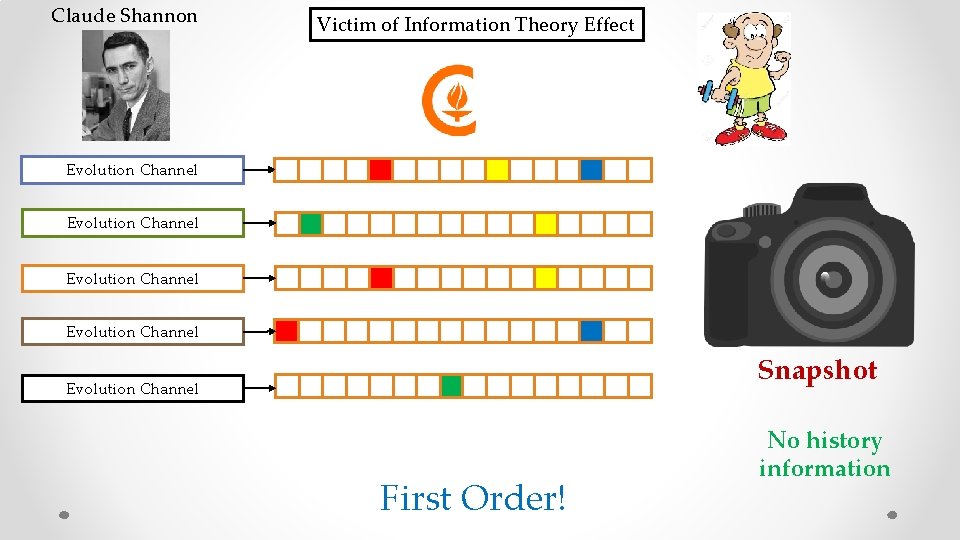
Claude Shannon Victim of Information Theory Effect Evolution Channel Snapshot Evolution Channel First Order! No history information

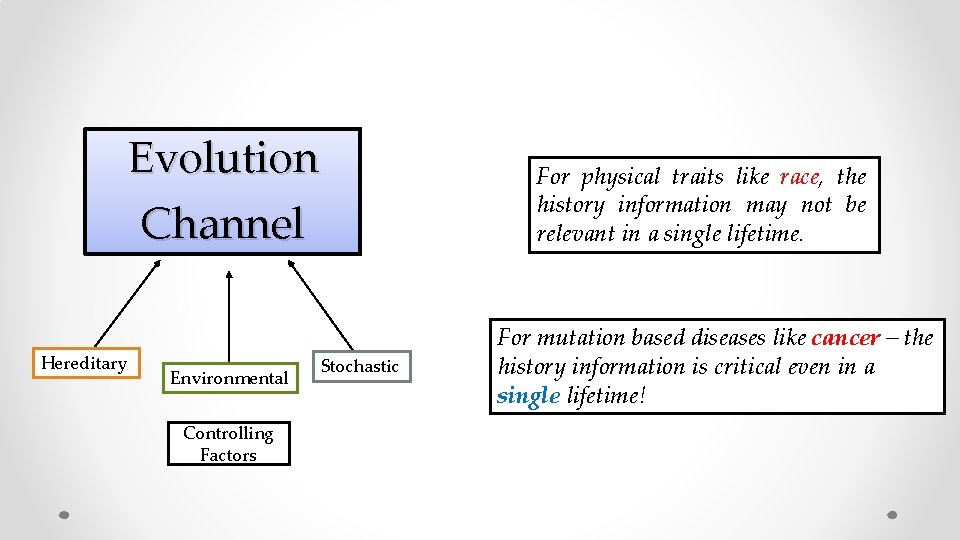
Evolution Channel Hereditary Environmental Controlling Factors Stochastic For physical traits like race, the history information may not be relevant in a single lifetime. For mutation based diseases like cancer – the history information is critical even in a single lifetime!

Can we use a single genome to gather information about the evolution channel?

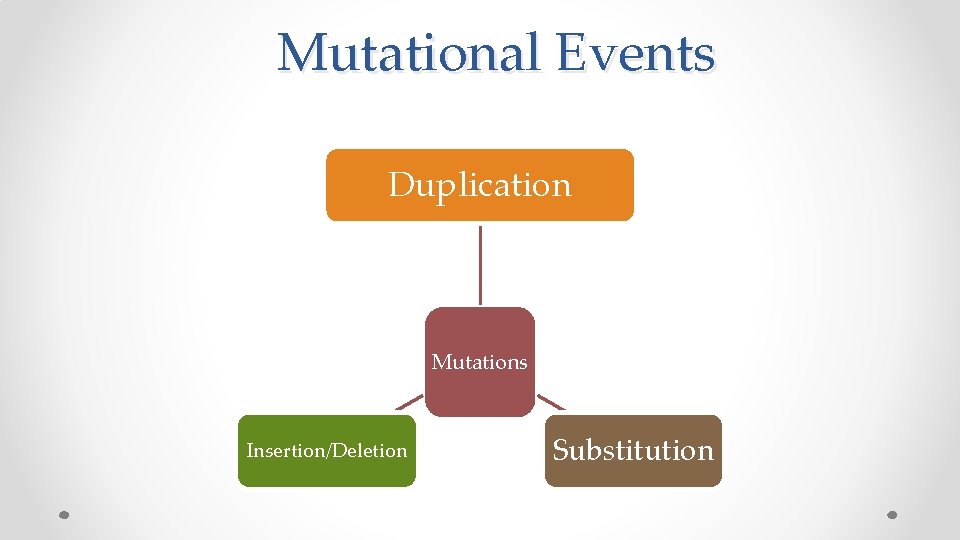
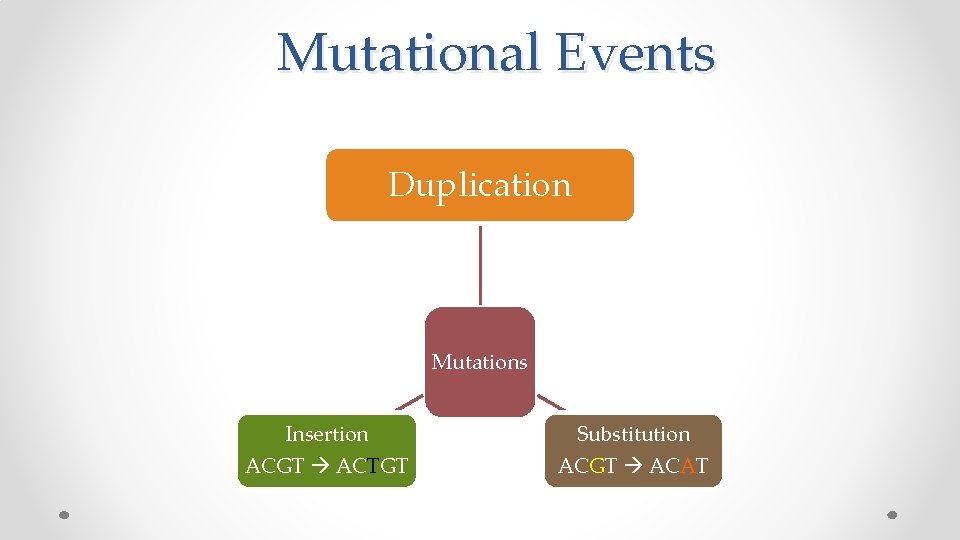
Mutational Events Duplication Mutations Insertion/Deletion Substitution

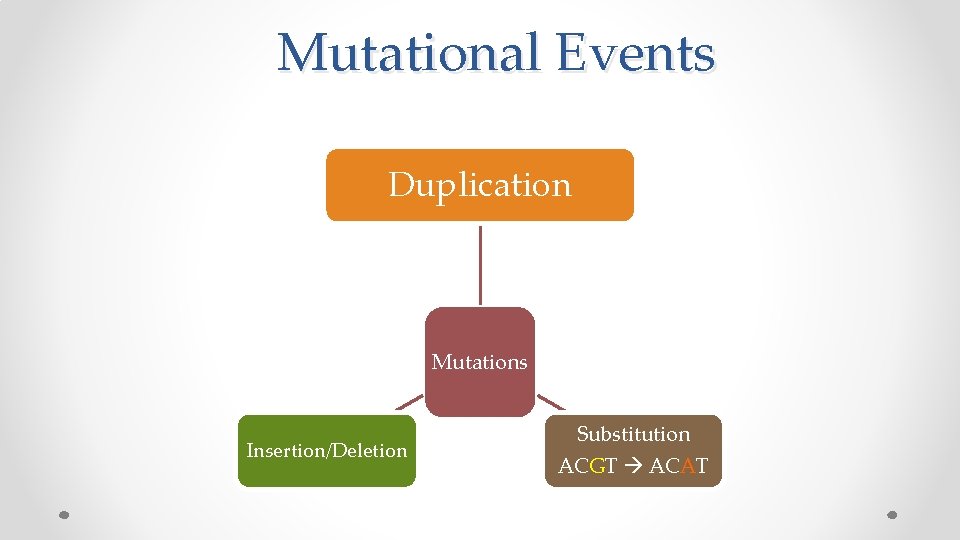
Mutational Events Duplication Mutations Insertion/Deletion Substitution ACGT ACAT

Mutational Events Duplication Mutations Insertion ACGT ACTGT Substitution ACGT ACAT

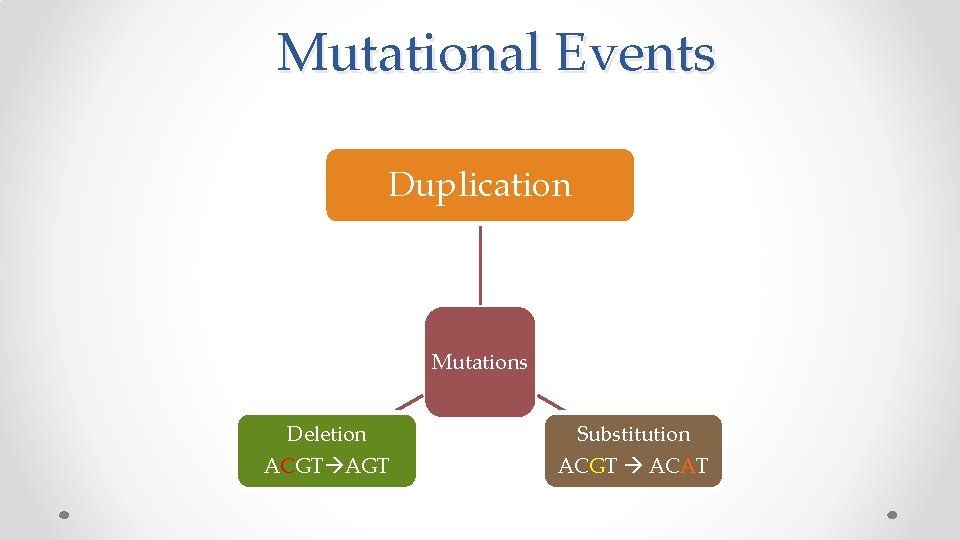
Mutational Events Duplication Mutations Deletion ACGT AGT Substitution ACGT ACAT

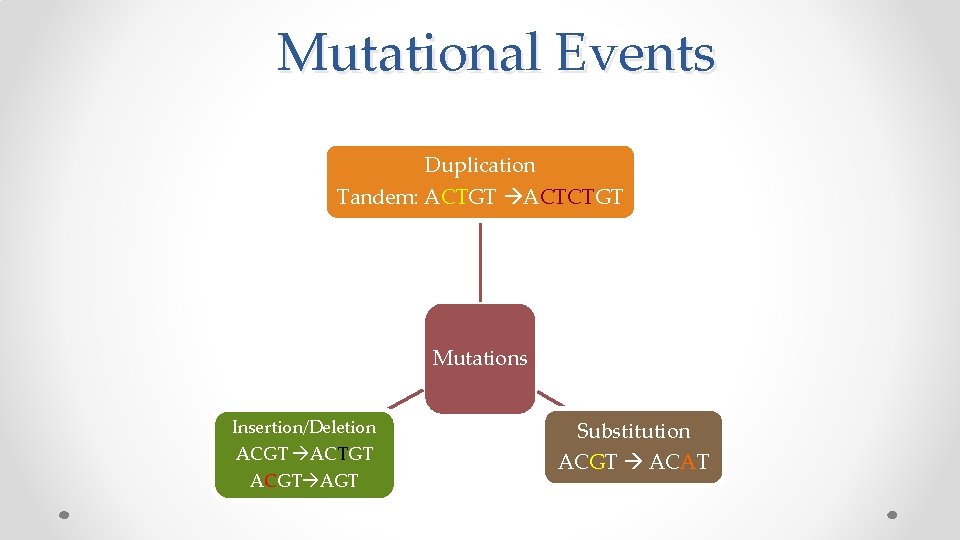
Mutational Events Duplication Tandem: ACTGT ACTCTGT Mutations Insertion/Deletion ACGT ACTGT ACGT AGT Substitution ACGT ACAT

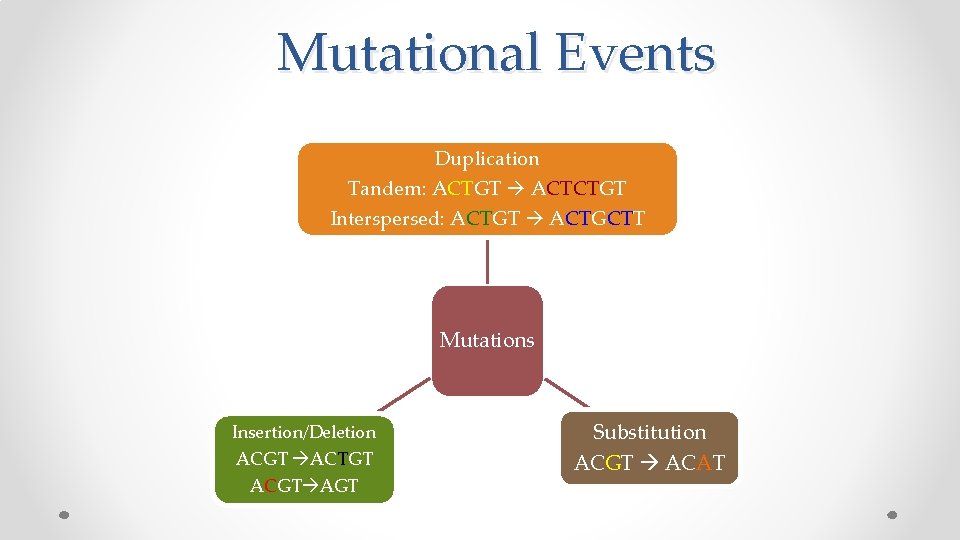
Mutational Events Duplication Tandem: ACTGT ACTCTGT Interspersed: ACTGT ACTGCTT Mutations Insertion/Deletion ACGT ACTGT ACGT AGT Substitution ACGT ACAT

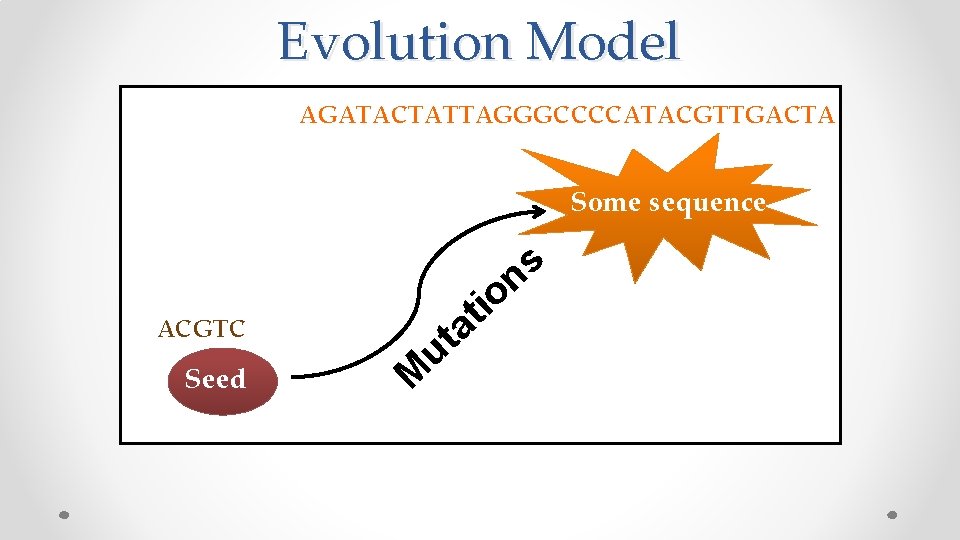
Evolution Model AGATACTATTAGGGCCCCATACGTTGACTA Some sequence s n it o a t u ACGTC Seed M

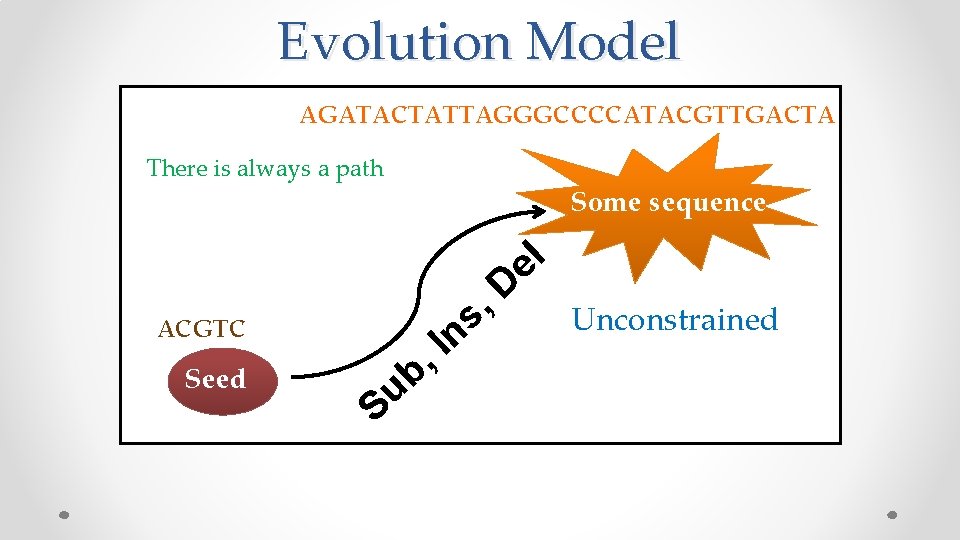
Evolution Model AGATACTATTAGGGCCCCATACGTTGACTA There is always a path Some sequence l e ACGTC Seed u S I , b s n D , Unconstrained

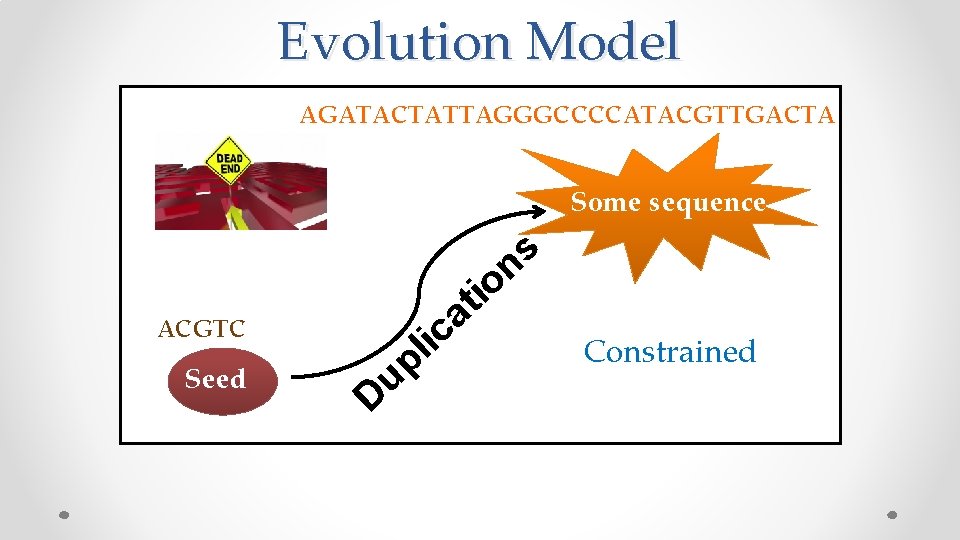
Evolution Model AGATACTATTAGGGCCCCATACGTTGACTA Some sequence a c ACGTC Seed it o u D i l p s n Constrained

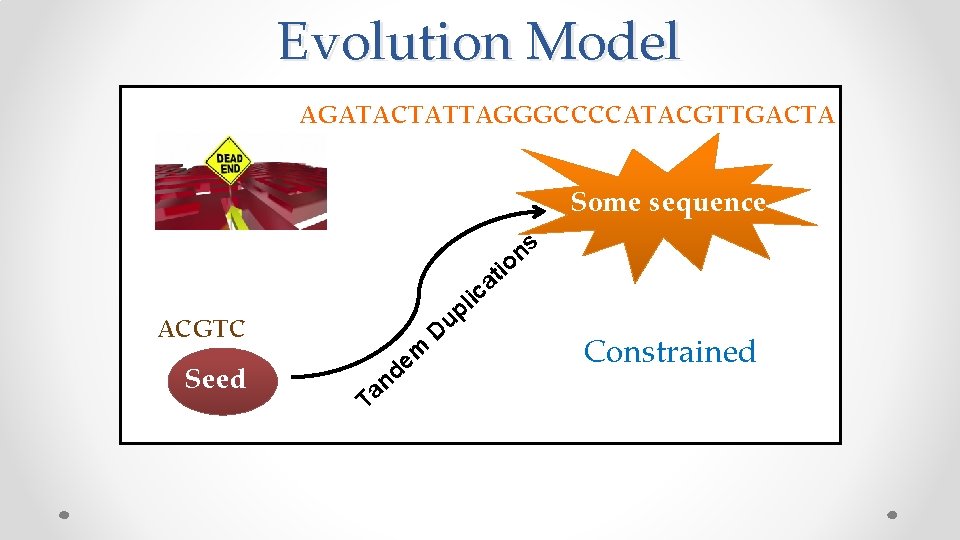
Evolution Model AGATACTATTAGGGCCCCATACGTTGACTA Some sequence s n o ti a c i ACGTC Seed l p u m e d n Ta D Constrained

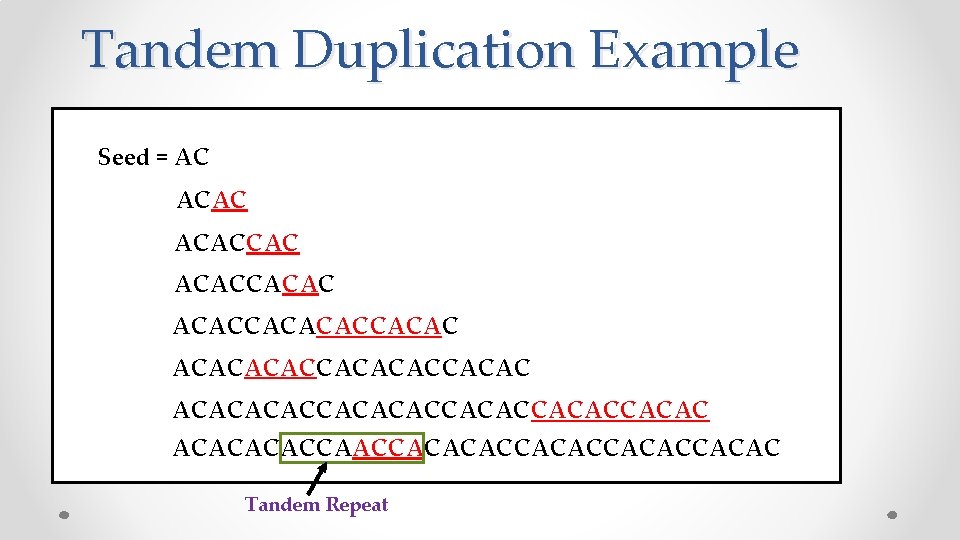
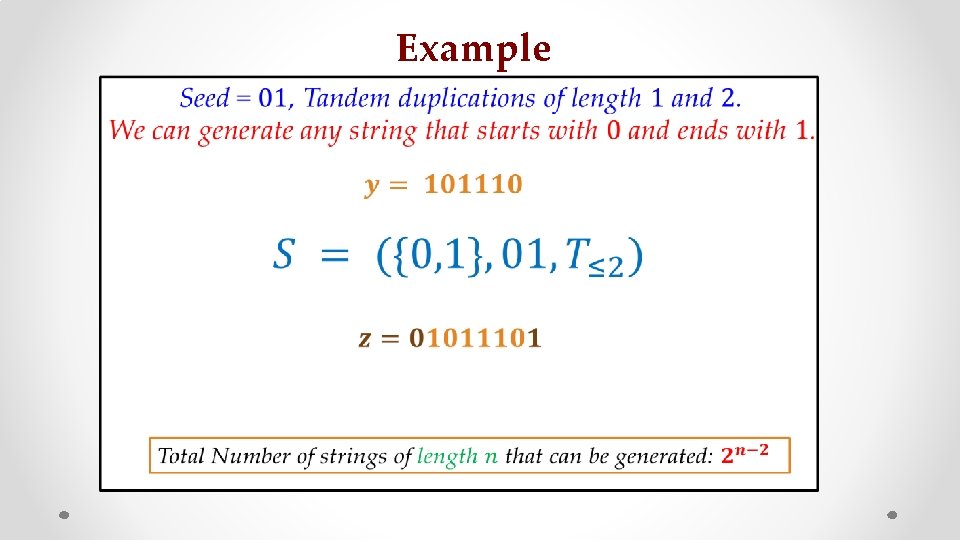
Tandem Duplication Example Seed = AC ACACCACACACCACACACACCACACACCACACCAACCACACCACAC Tandem Repeat

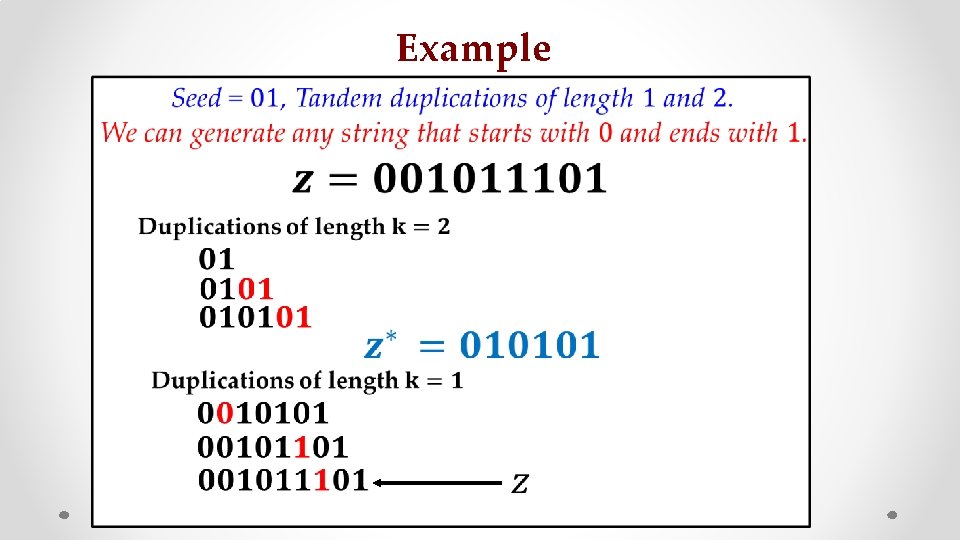
Example • z s

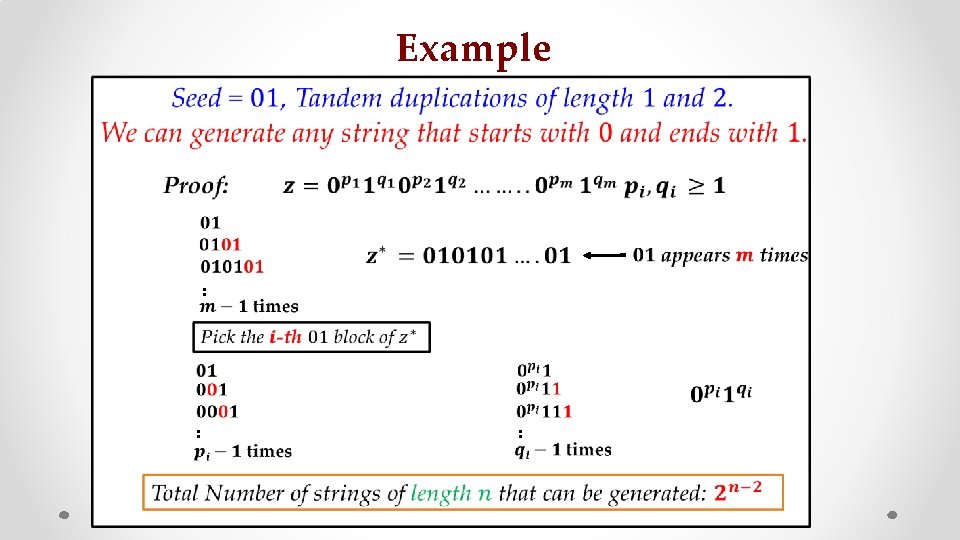
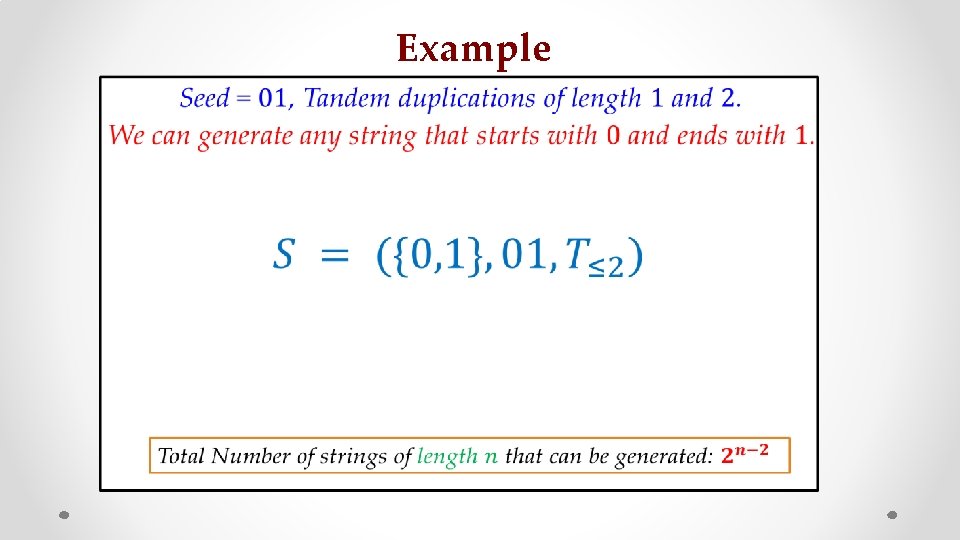
Example • : z : s :
![Definitions F Farnoud M Schwartz J Bruck ISIT 14 Definitions • [F. Farnoud, M. Schwartz, J. Bruck, ISIT’ 14]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-23.jpg)
Definitions • [F. Farnoud, M. Schwartz, J. Bruck, ISIT’ 14]

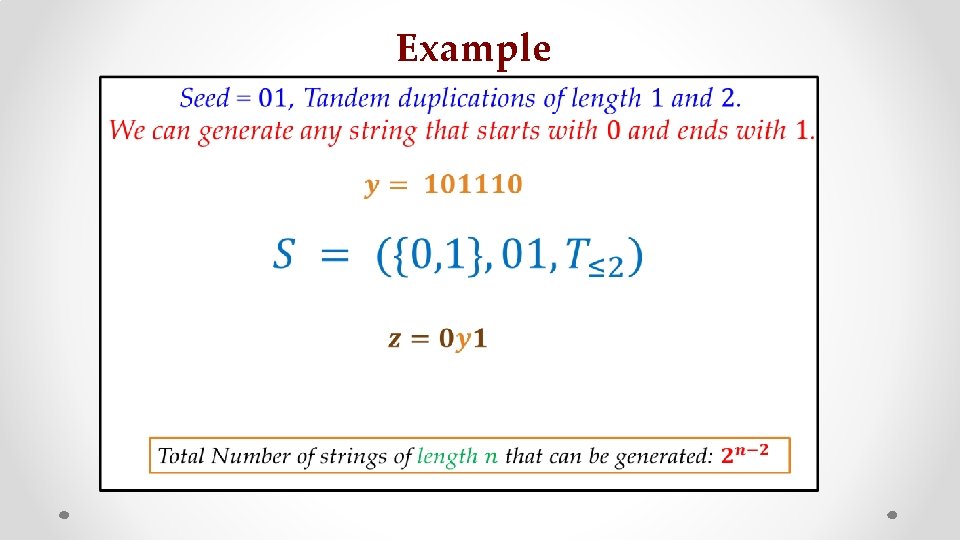
Example • z s

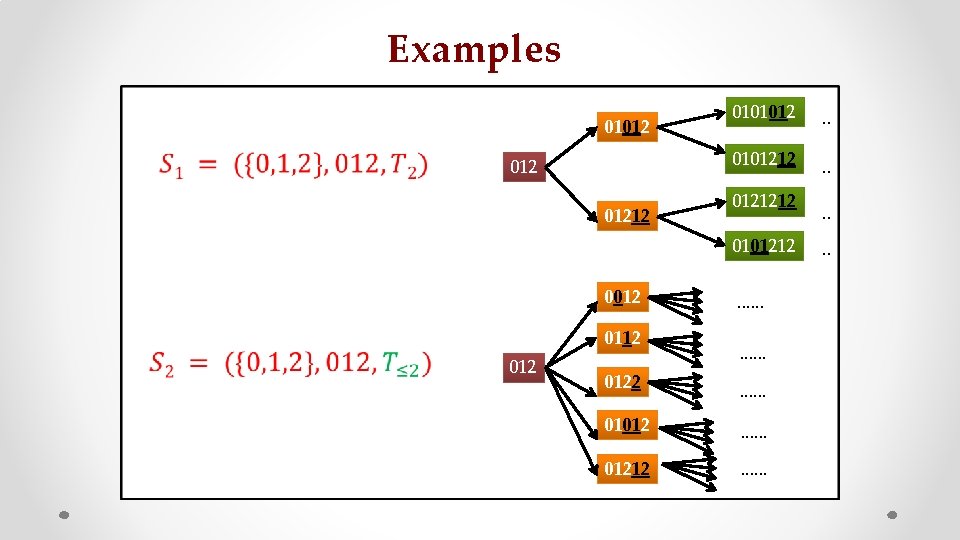
Examples 01012 01212 0101012 . . 0101212 . . 0121212 0101212 0012 0112 0122 . . . . 01012 . . . 01212 . .

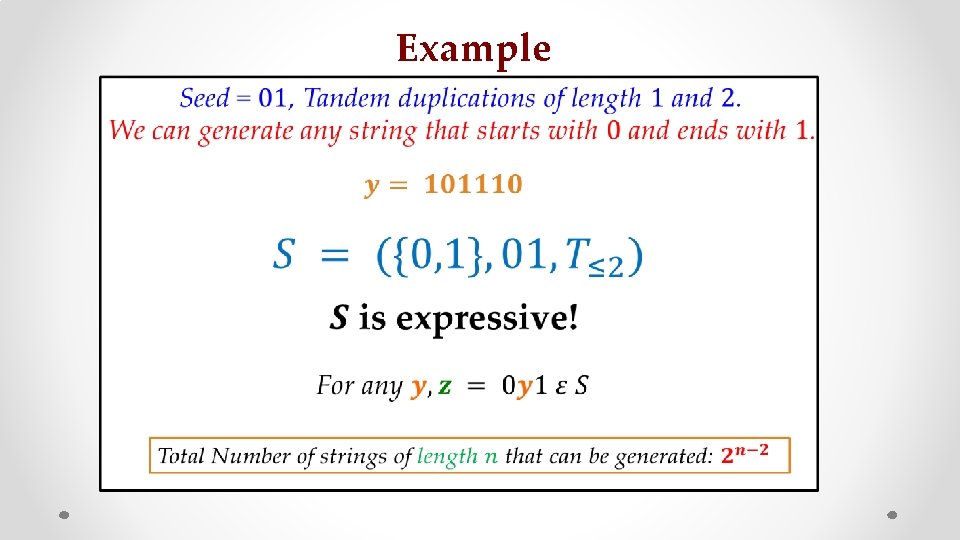
Diversity S. Jain, F. Farnoud, J. Bruck, ‘’Capacity and Expressiveness of Genomic Tandem Duplication” IEEE IT 2017 Tandem Duplications seed

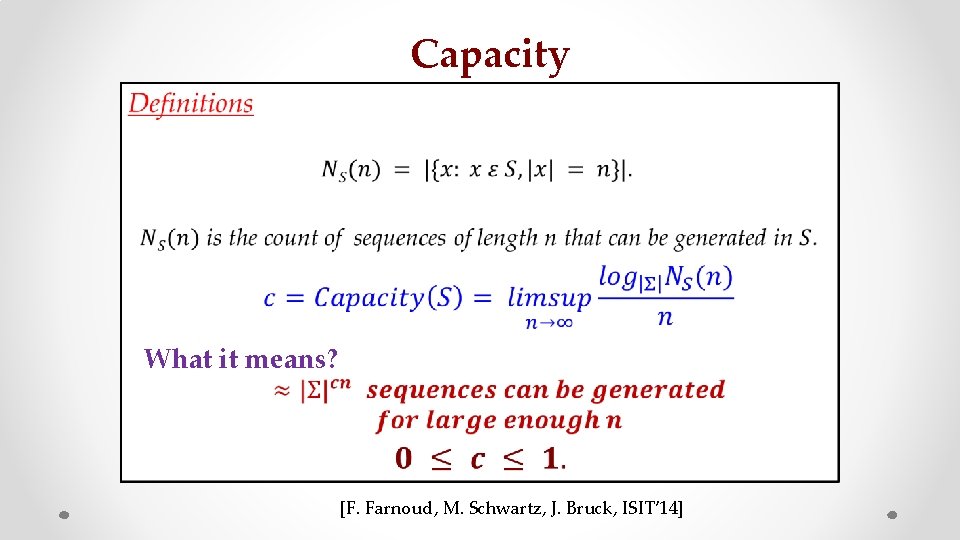
Capacity • z What it means? s [F. Farnoud, M. Schwartz, J. Bruck, ISIT’ 14]

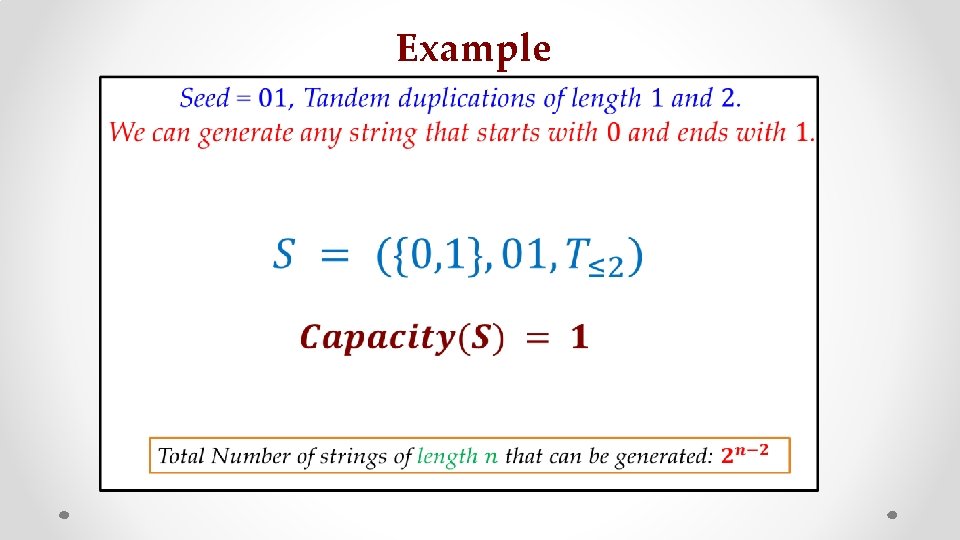
Example • s z

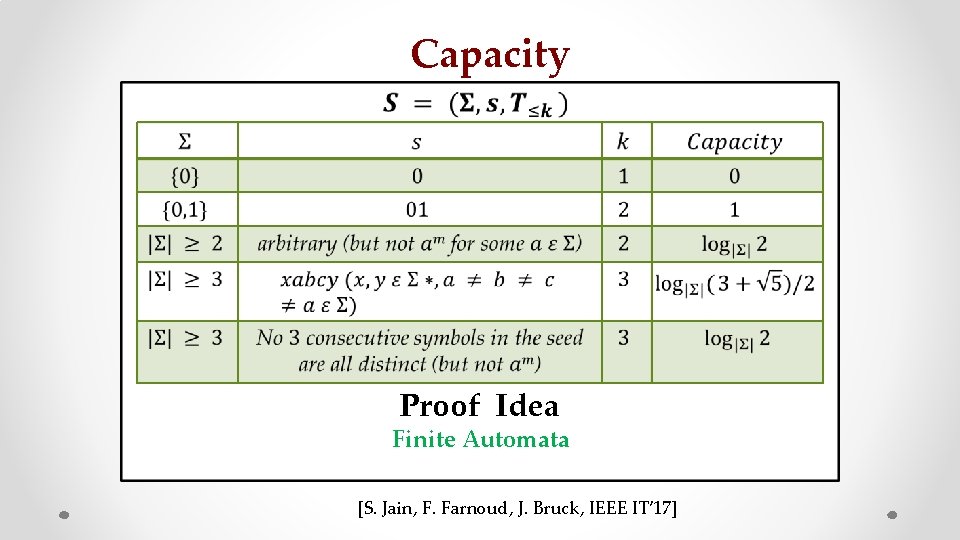
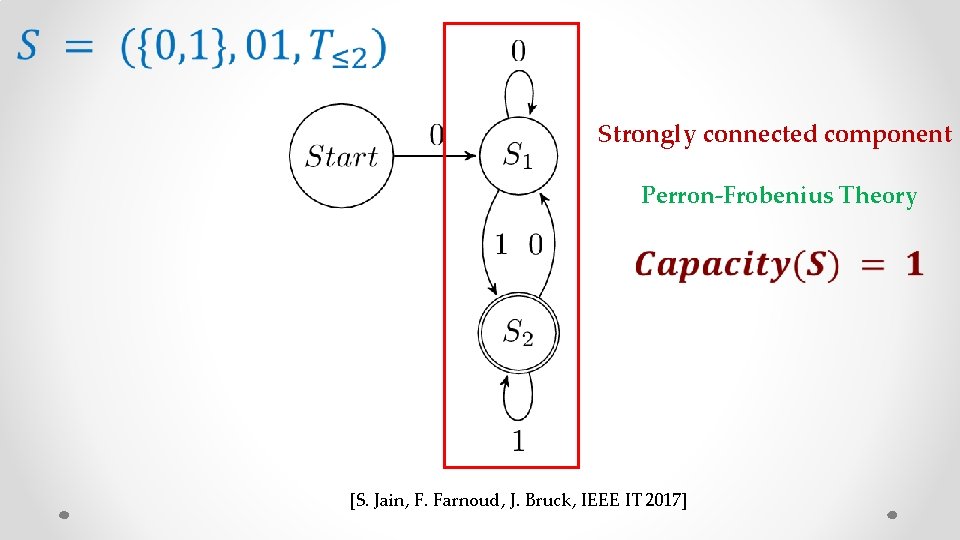
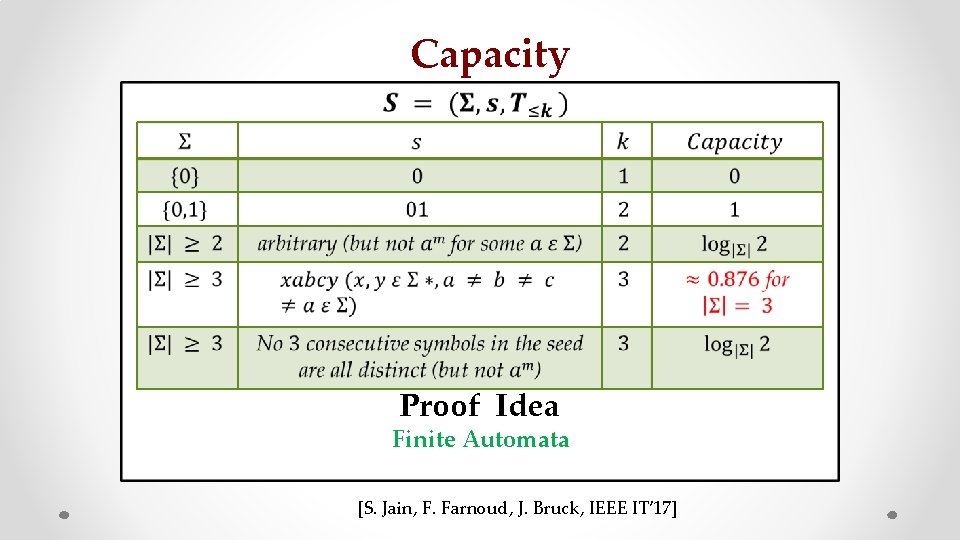
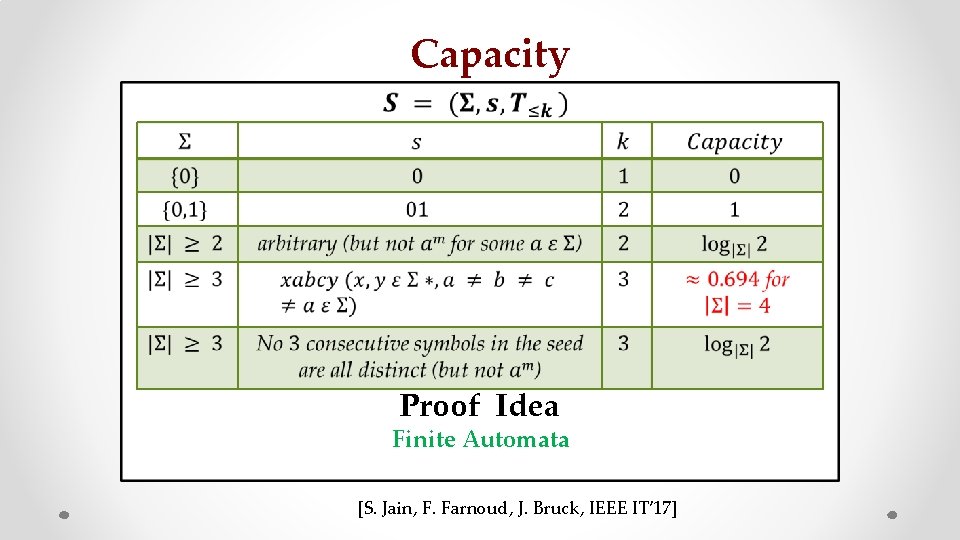
Capacity • z Proof Idea Finite Automata [S. Jain, F. Farnoud, J. Bruck, IEEE IT’ 17]

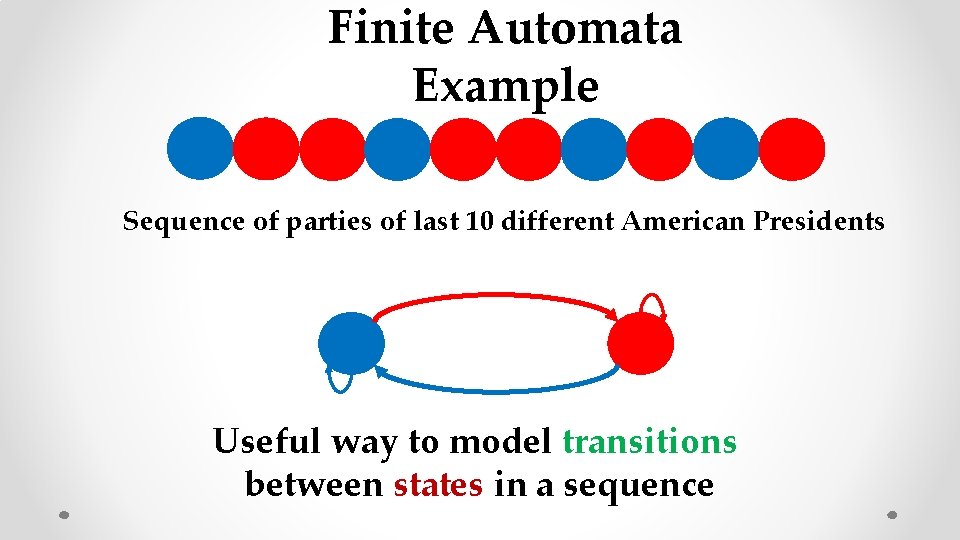
Finite Automata Example Sequence of parties of last 10 different American Presidents Useful way to model transitions between states in a sequence

Strongly connected component Perron-Frobenius Theory [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]

Capacity • z Proof Idea Finite Automata [S. Jain, F. Farnoud, J. Bruck, IEEE IT’ 17]

Capacity • z Proof Idea Finite Automata [S. Jain, F. Farnoud, J. Bruck, IEEE IT’ 17]
![S Jain F Farnoud J Bruck IEEE IT 2017 [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-34.jpg)
[S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]
![Arbitrary Seed S Jain F Farnoud J Bruck IEEE IT 2017 Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-35.jpg)
Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]
![Arbitrary Seed S Jain F Farnoud J Bruck IEEE IT 2017 Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-36.jpg)
Arbitrary Seed [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]

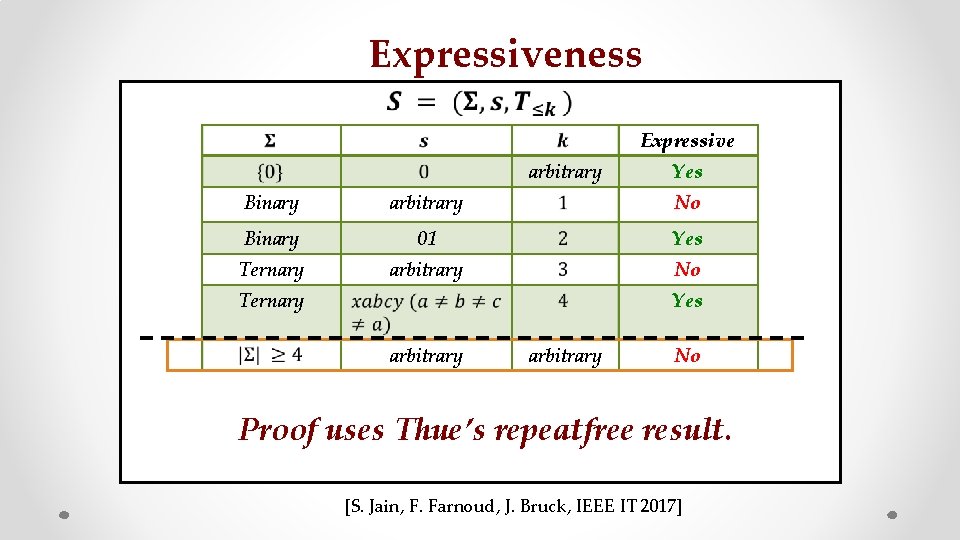
Expressiveness Seed

Expressiveness Seed

Example • s z

Example • s z

Example • s z

Example • z s

Expressiveness Expressive arbitrary Yes Binary arbitrary No Binary 01 Yes Ternary arbitrary No Ternary Yes arbitrary z No Proof uses Thue’s repeatfree result. [S. Jain, F. Farnoud, J. Bruck, IEEE IT 2017]

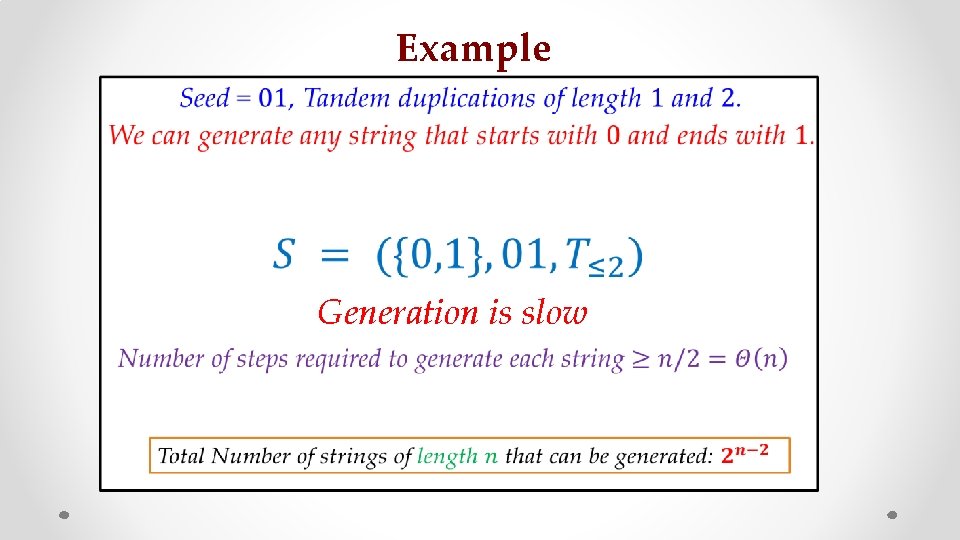
Example • Generation is slow s z

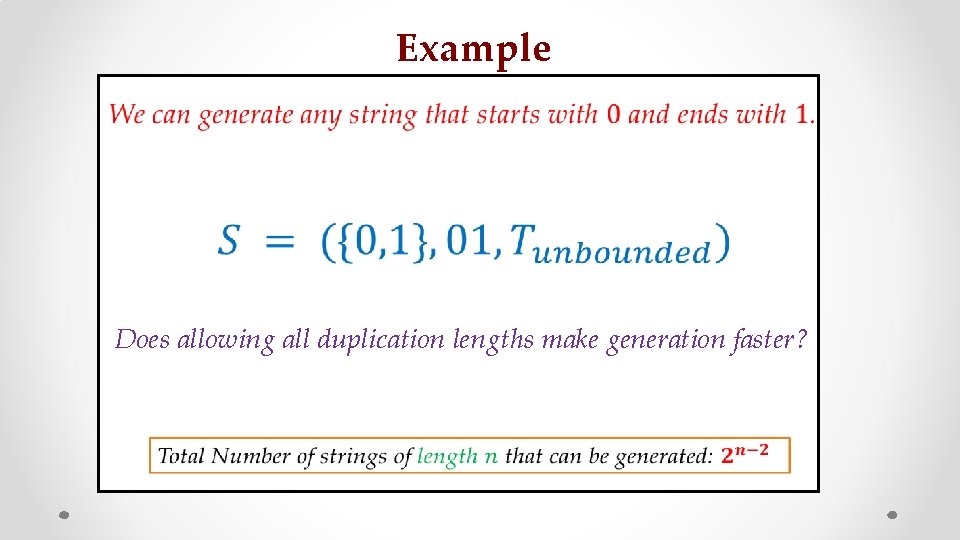
Example z faster? Does allowing all duplication lengths make generation s

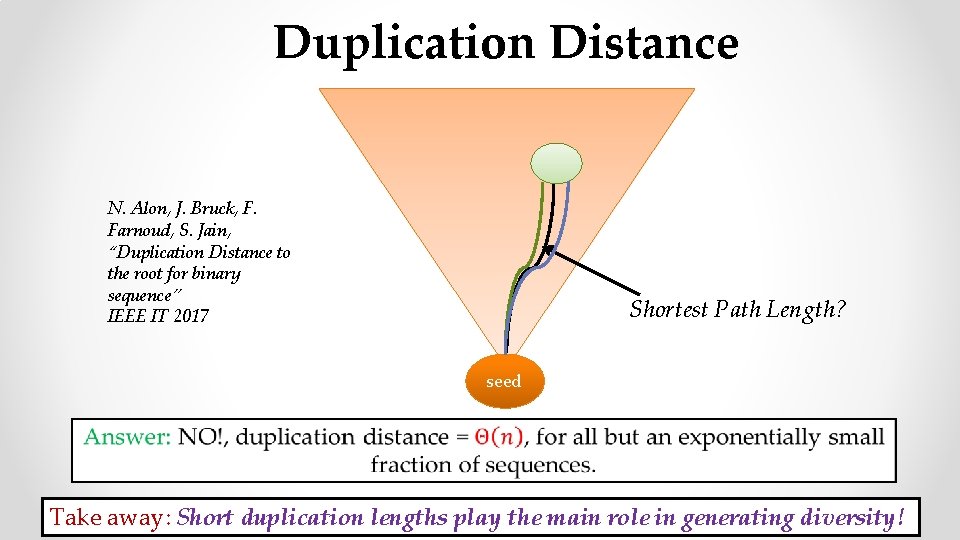
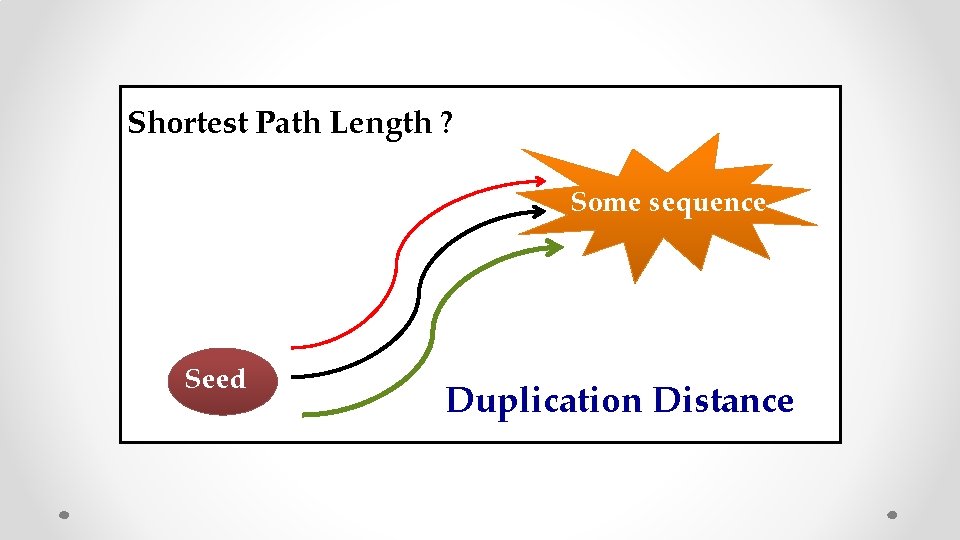
Duplication Distance N. Alon, J. Bruck, F. Farnoud, S. Jain, “Duplication Distance to the root for binary sequence” IEEE IT 2017 Shortest Path Length? seed Take away: Short duplication lengths play the main role in generating diversity!

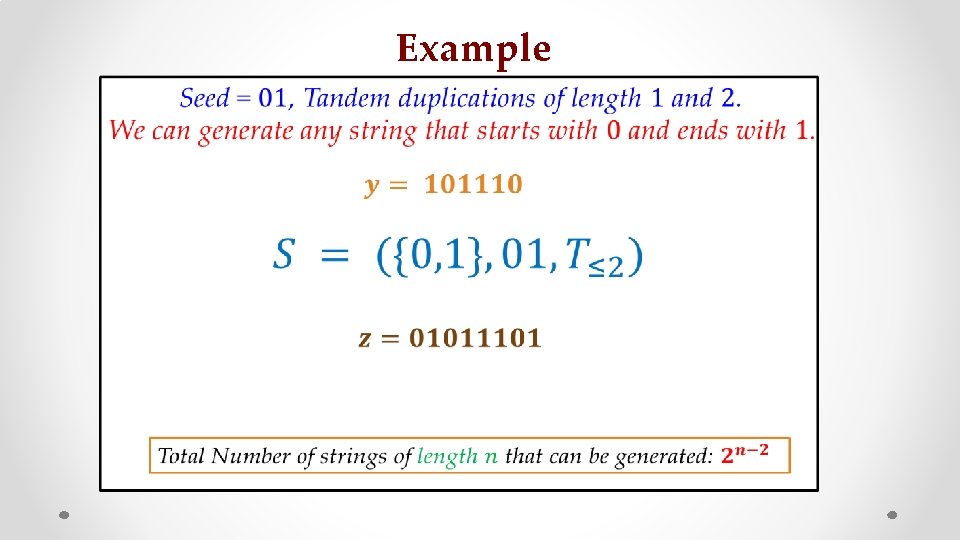
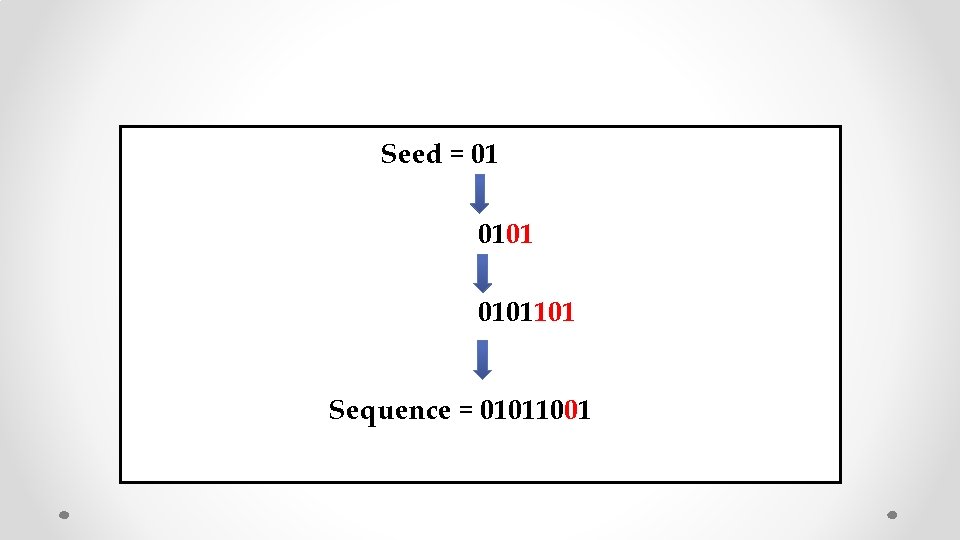
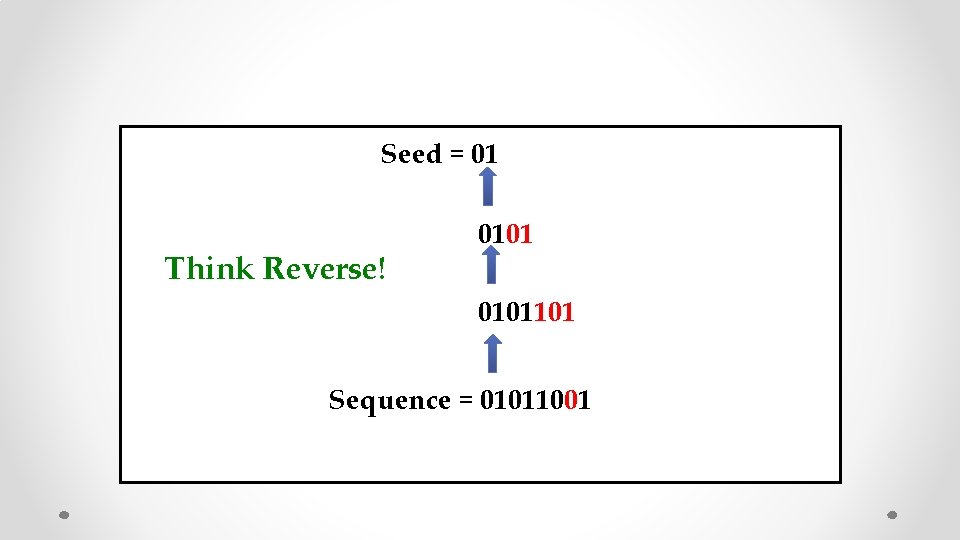
Seed = 01 0101101 Sequence = 01011001

Seed = 01 Think Reverse! 0101101 Sequence = 01011001

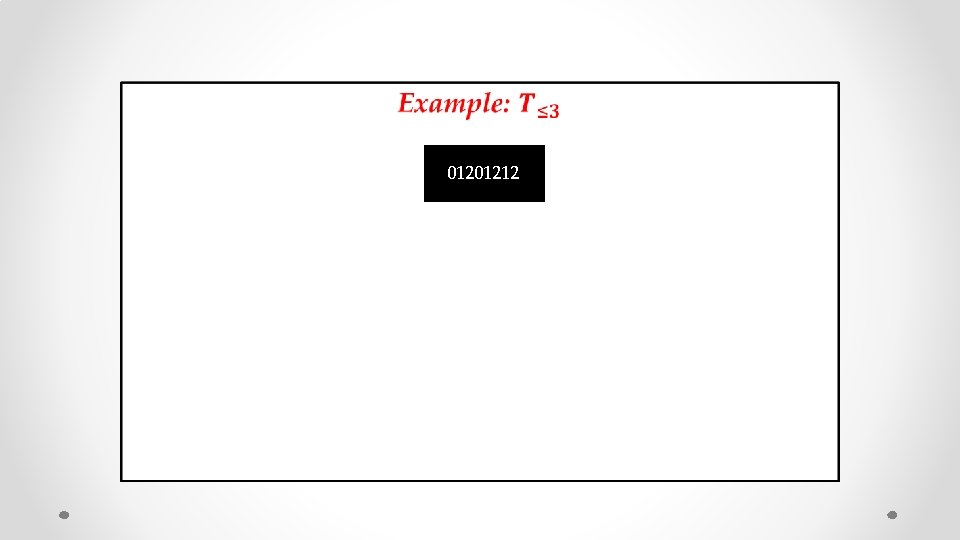
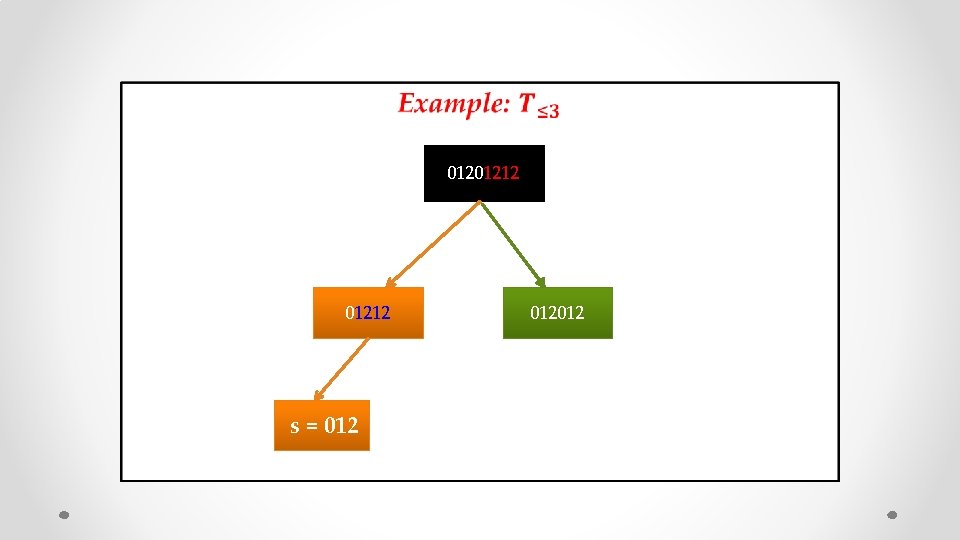
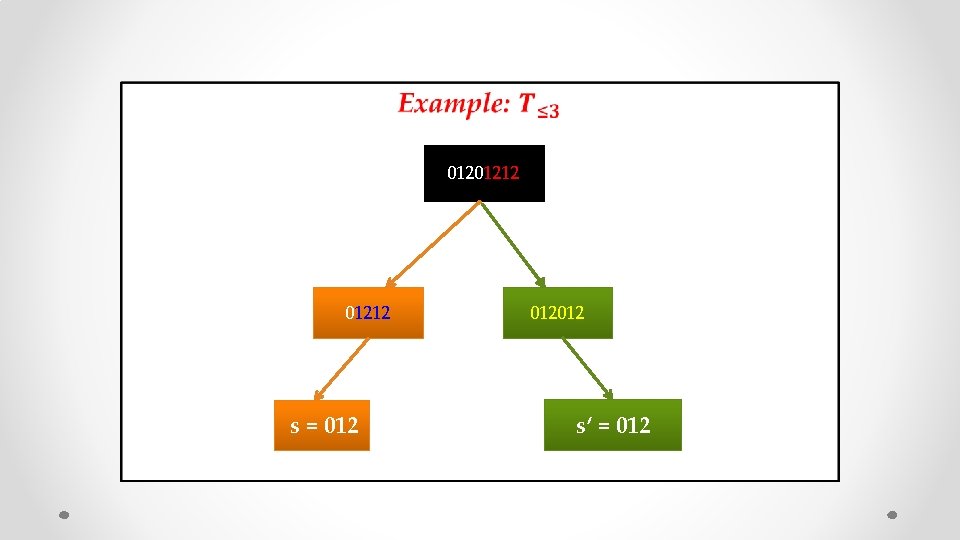
• 01201212

• 01201212

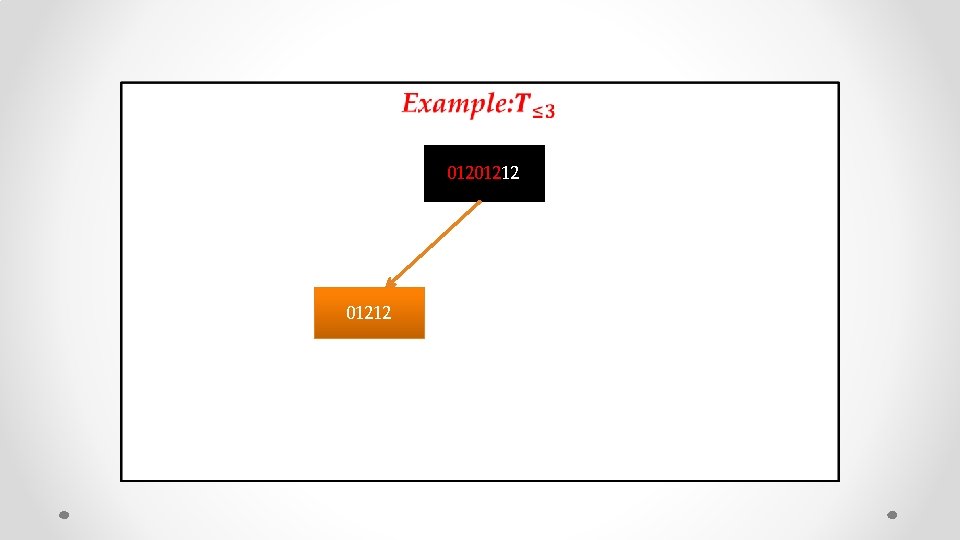
• 01201212 s = 012

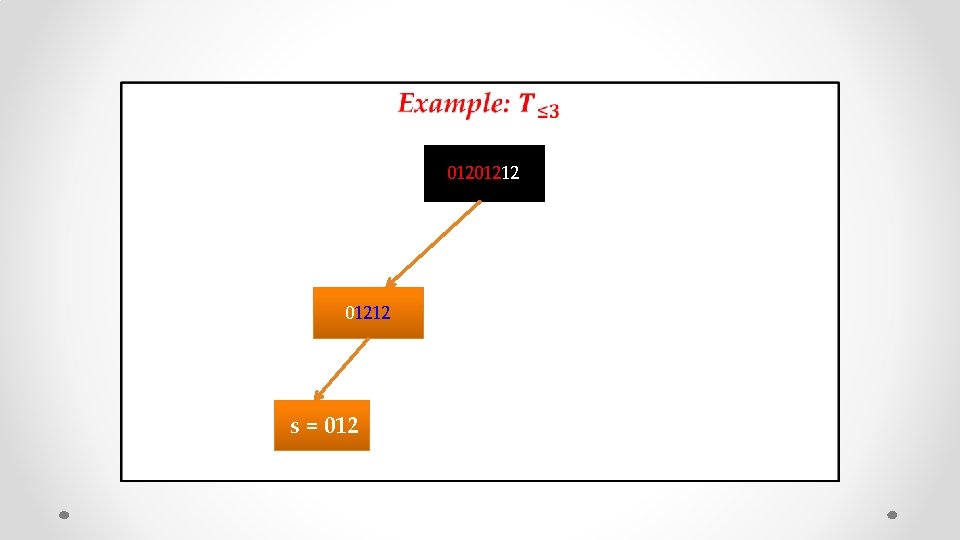
• 01201212 s = 012012

• 01201212 s = 012012 s’ = 012

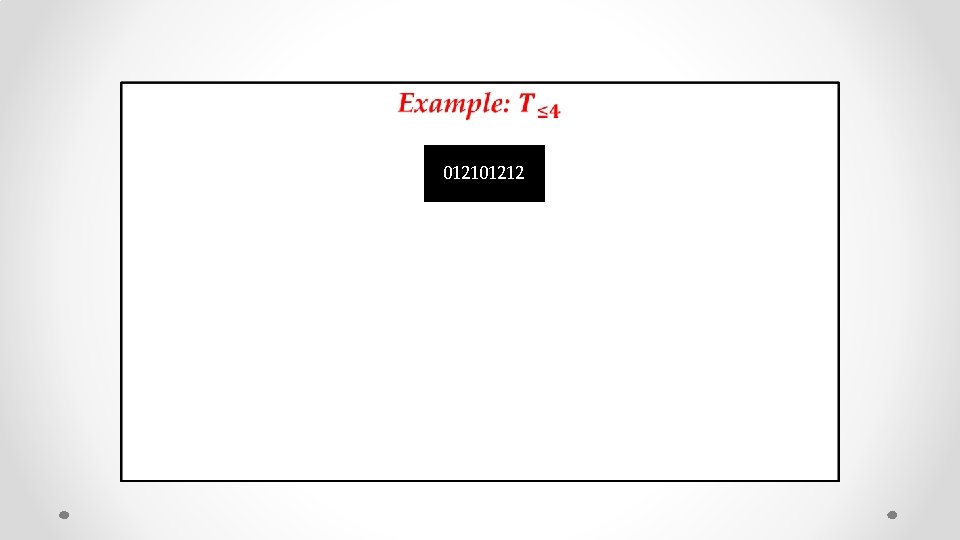
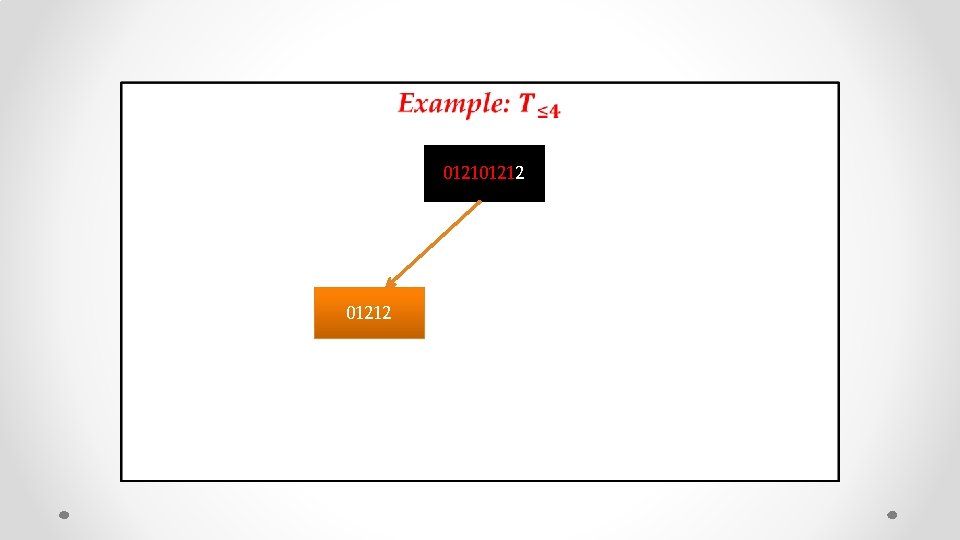
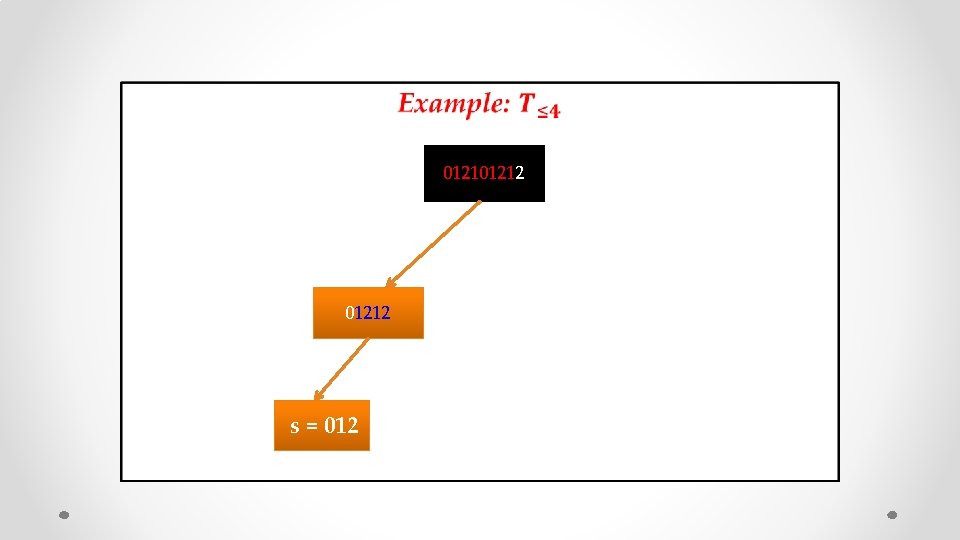
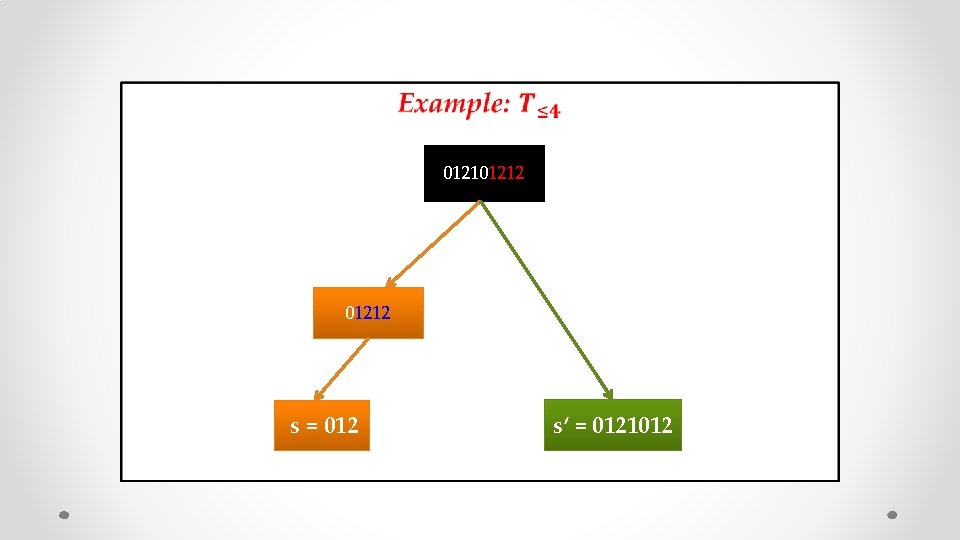
• 01212

• 01212

• 01212 s = 012

• 01212 s = 012 s’ = 0121012

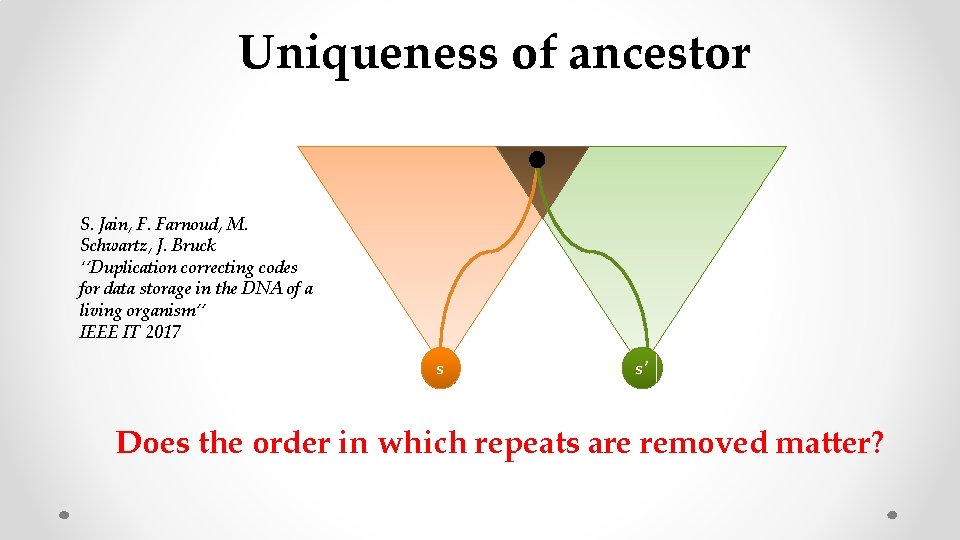
Uniqueness of ancestor S. Jain, F. Farnoud, M. Schwartz, J. Bruck ‘’Duplication correcting codes for data storage in the DNA of a living organism’’ IEEE IT 2017 s s’ Does the order in which repeats are removed matter?

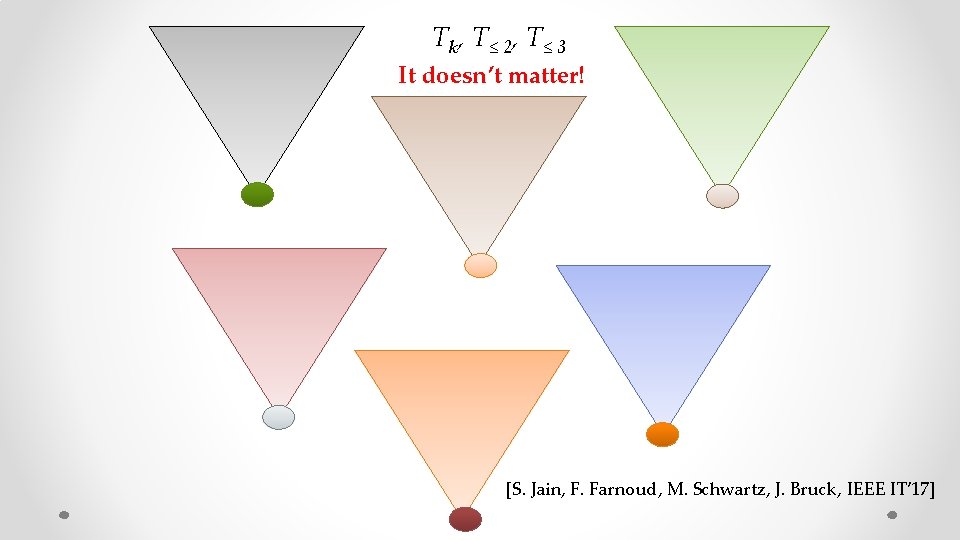
Tk, T≤ 2, T≤ 3 It doesn’t matter! [S. Jain, F. Farnoud, M. Schwartz, J. Bruck, IEEE IT’ 17]

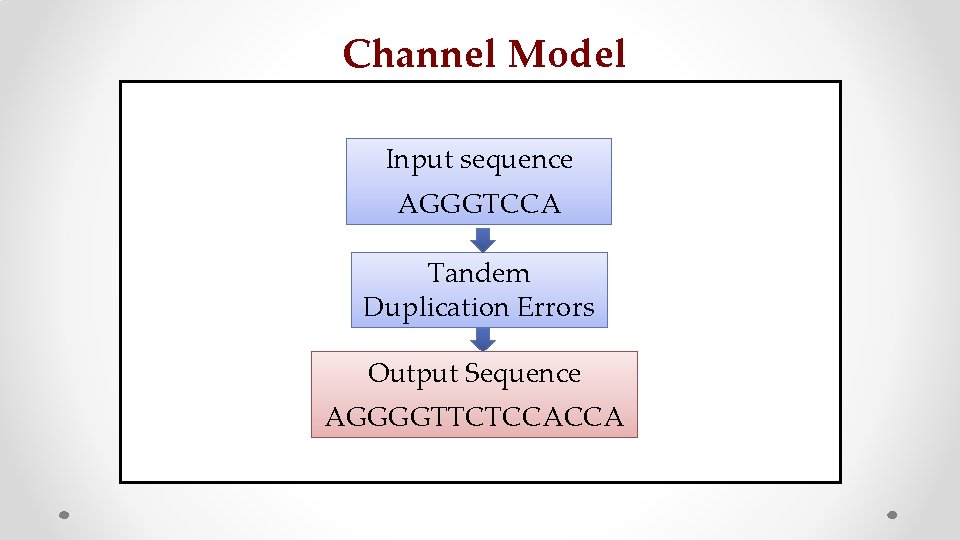
Channel Model Input sequence AGGGTCCA Tandem Duplication Errors Output Sequence AGGGGTTCTCCACCA

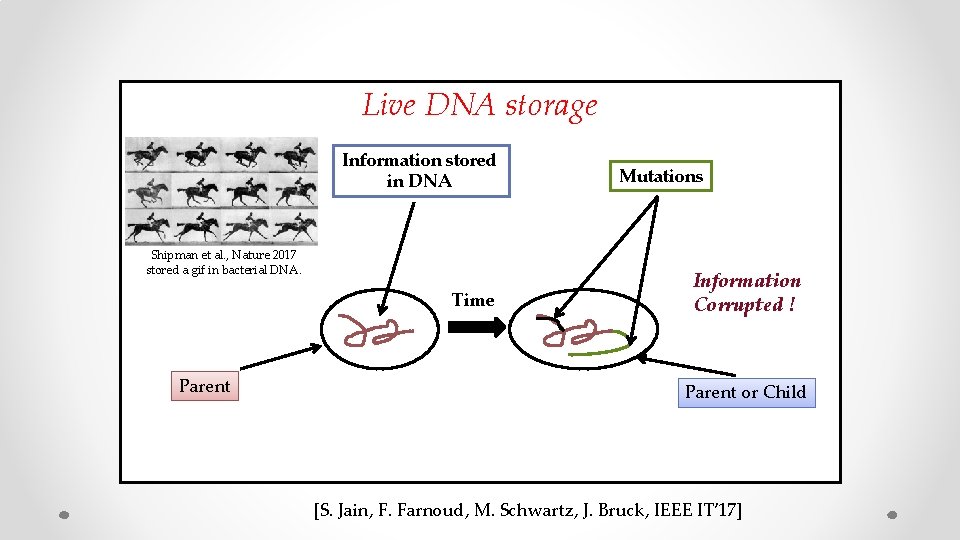
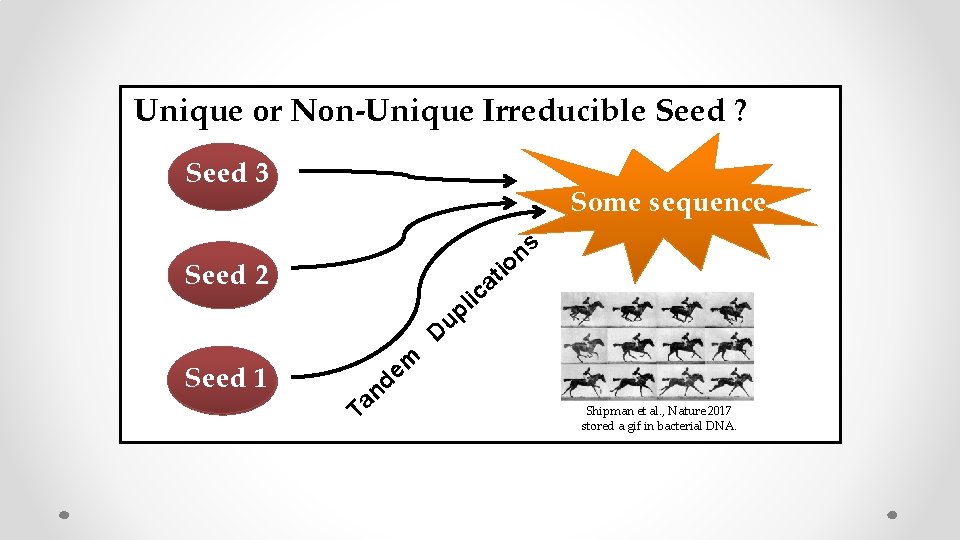
Live DNA storage Information stored in DNA Shipman et al. , Nature 2017 stored a gif in bacterial DNA. Time Parent Mutations Information Corrupted ! Parent or Child [S. Jain, F. Farnoud, M. Schwartz, J. Bruck, IEEE IT’ 17]

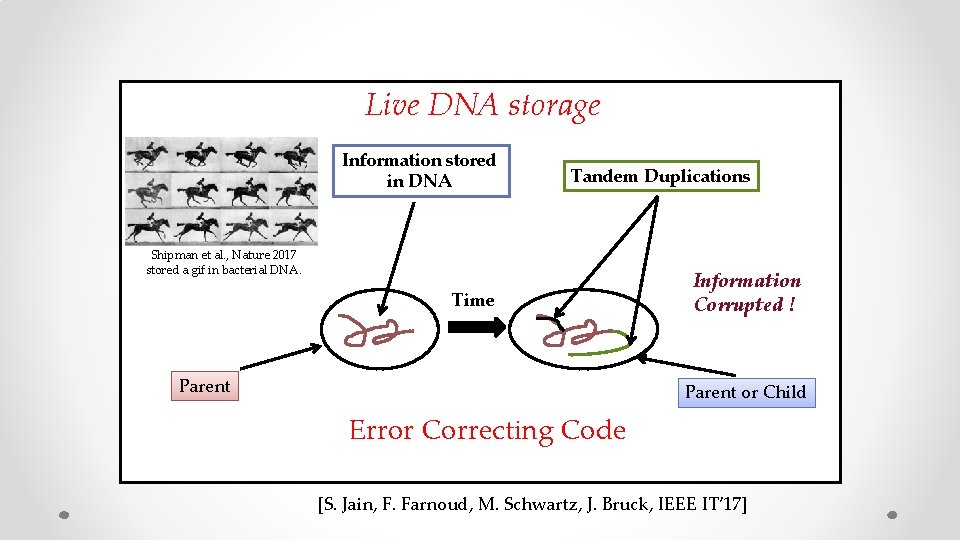
Live DNA storage Information stored in DNA Tandem Duplications Shipman et al. , Nature 2017 stored a gif in bacterial DNA. Time Parent Information Corrupted ! Parent or Child Error Correcting Code [S. Jain, F. Farnoud, M. Schwartz, J. Bruck, IEEE IT’ 17]

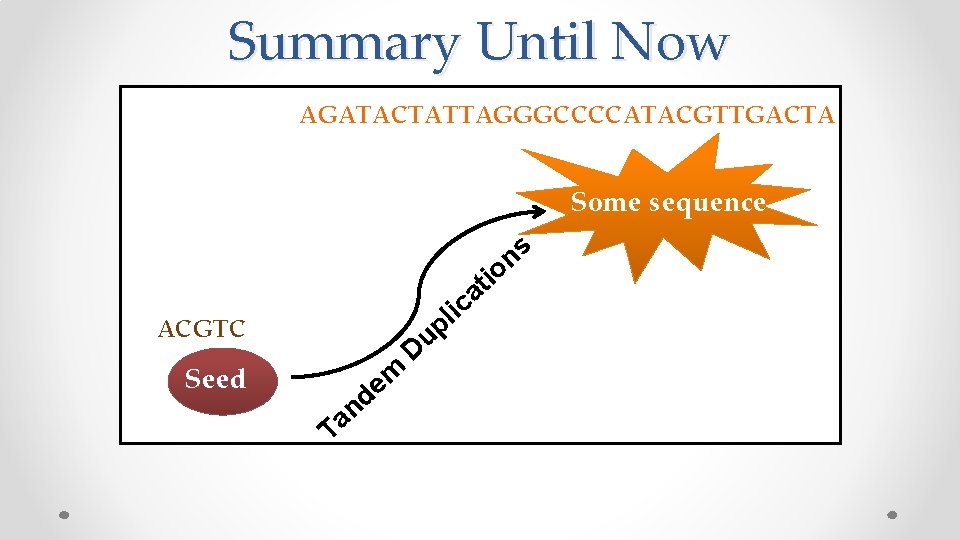
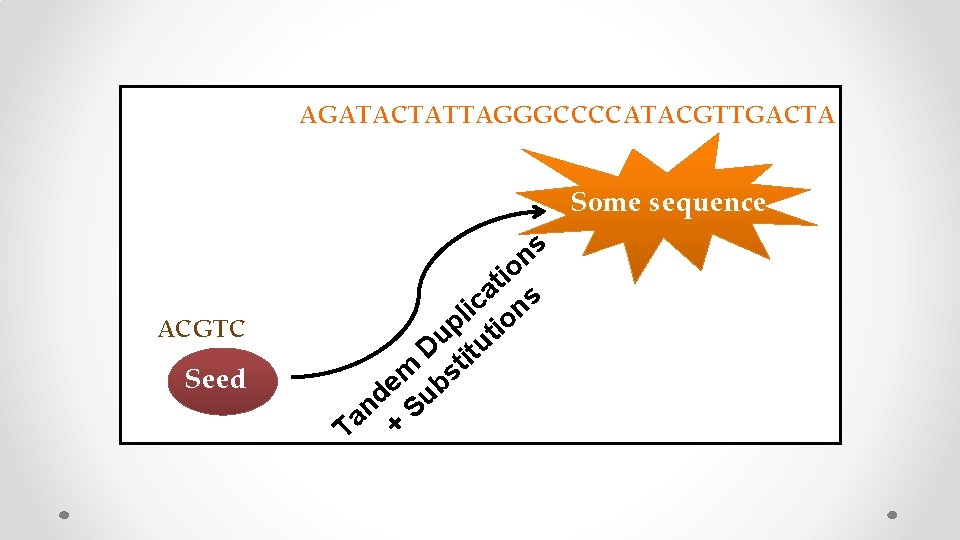
Summary Until Now AGATACTATTAGGGCCCCATACGTTGACTA Some sequence s n tio a c li ACGTC Seed n a T m e d p u D

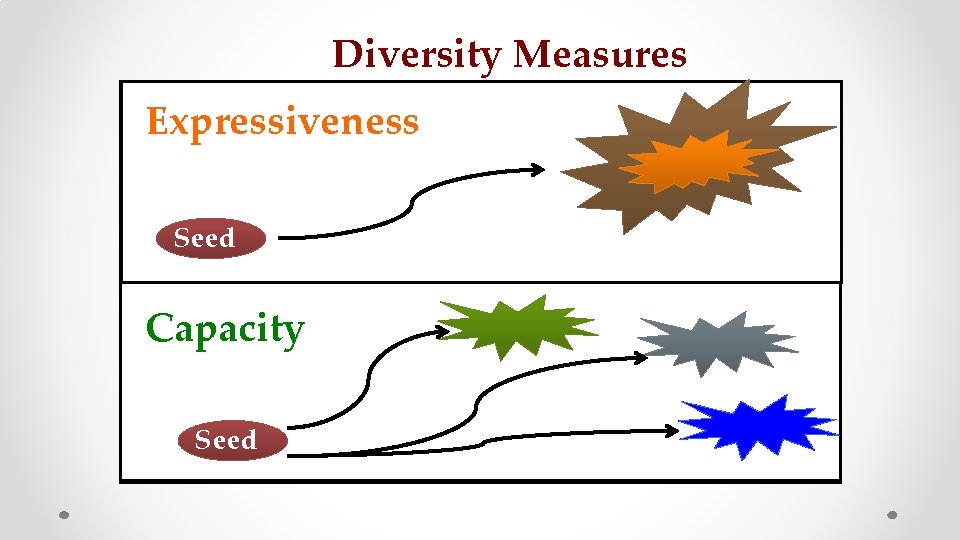
Diversity Measures Expressiveness Seed Capacity Seed

Shortest Path Length ? Some sequence Seed Duplication Distance

Unique or Non-Unique Irreducible Seed ? Seed 3 Some sequence Seed 2 ti a ic l up s n o D Seed 1 Seed Ta m e nd Bonnet et al. , 2012 Shipman et al. , Nature 2017 stored a gif in bacterial DNA.

AGATACTATTAGGGCCCCATACGTTGACTA Some sequence ACGTC Seed s n io t a s c i on l p ti u D titu m bs e d u n S a T +

Moving to real DNA data

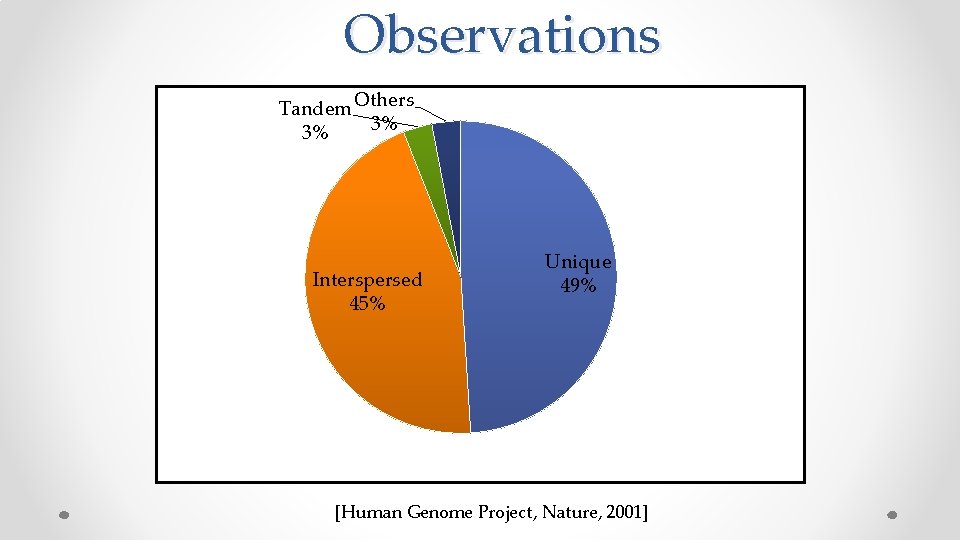
Observations Tandem Others 3% 3% Interspersed 45% Unique 49% [Human Genome Project, Nature, 2001]

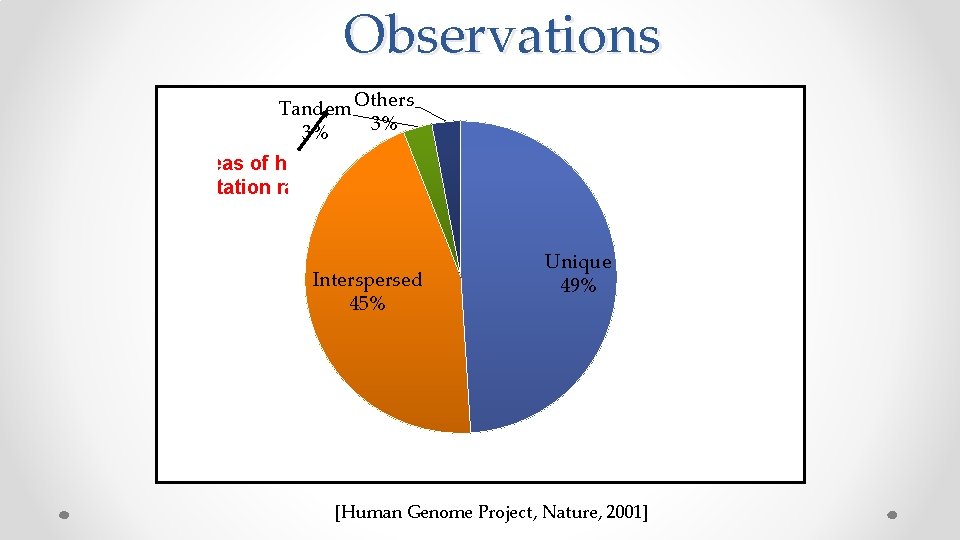
Observations Tandem Others 3% 3% Areas of high mutation rates Interspersed 45% Unique 49% [Human Genome Project, Nature, 2001]

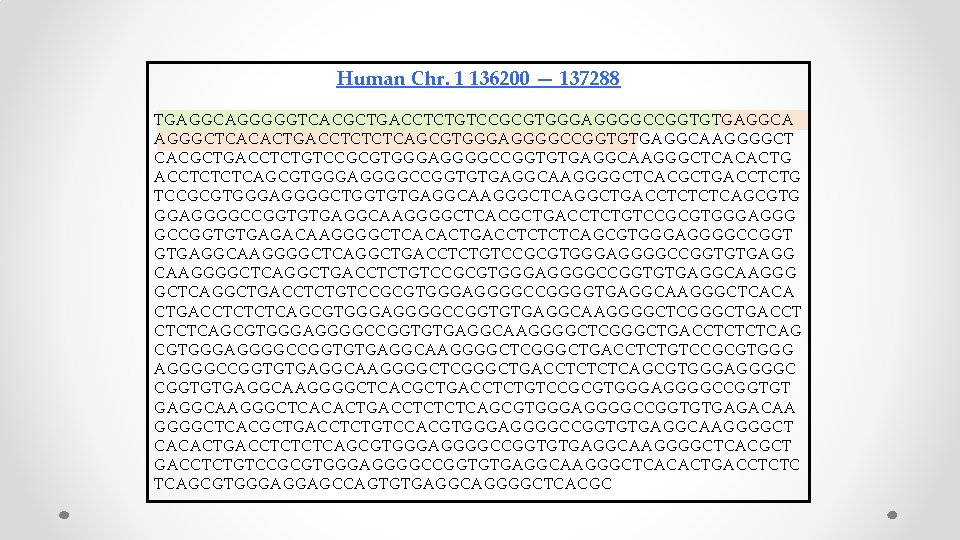
Human Chr. 1 136200 — 137288 TGAGGCAGGGGGTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTGTGAGGCA AGGGCTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTGTGAGGCAAGGGGCT CACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTGTGAGGCAAGGGCTCACACTG ACCTCTCTCAGCGTGGGAGGGGCCGGTGTGAGGCAAGGGGCTCACGCTGACCTCTG TCCGCGTGGGAGGGGCTGGTGTGAGGCAAGGGCTCAGGCTGACCTCTCTCAGCGTG GGAGGGGCCGGTGTGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGG GCCGGTGTGAGACAAGGGGCTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGT GTGAGGCAAGGGGCTCAGGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTGTGAGGCAAGGG GCTCAGGCTGACCTCTGTCCGCGTGGGAGGGGCCGGGGTGAGGCAAGGGCTCACA CTGACCTCTCTCAGCGTGGGAGGGGCCGGTGTGAGGCAAGGGGCTCGGGCTGACCTCTCTCAG CGTGGGAGGGGCCGGTGTGAGGCAAGGGGCTCGGGCTGACCTCTGTCCGCGTGGG AGGGGCCGGTGTGAGGCAAGGGGCTCGGGCTGACCTCTCTCAGCGTGGGAGGGGC CGGTGTGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTGT GAGGCAAGGGCTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTGTGAGACAA GGGGCTCACGCTGACCTCTGTCCACGTGGGAGGGGCCGGTGTGAGGCAAGGGGCT CACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTGTGAGGCAAGGGGCTCACGCT GACCTCTGTCCGCGTGGGAGGGGCCGGTGTGAGGCAAGGGCTCACACTGACCTCTC TCAGCGTGGGAGGAGCCAGTGTGAGGCAGGGGCTCACGC

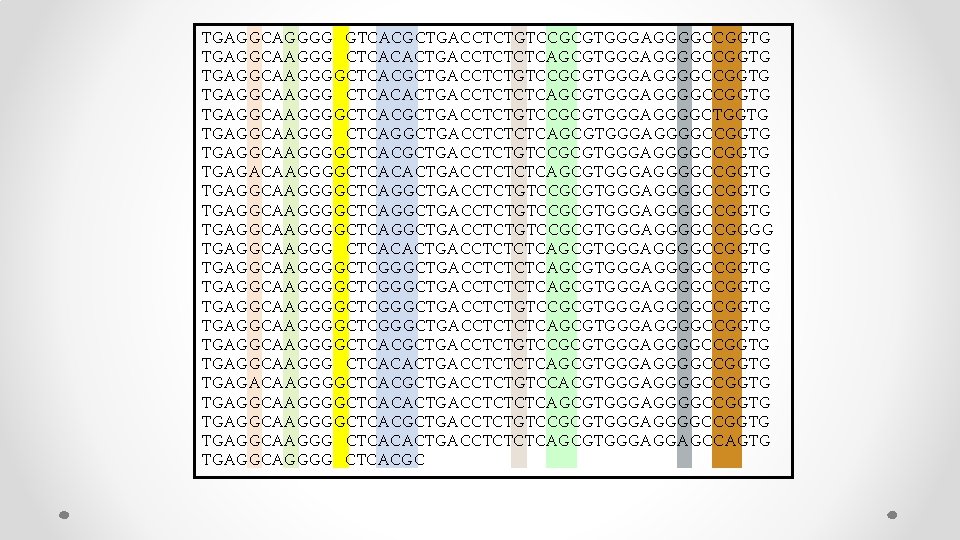
TGAGGCAGGGG GTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGG CTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGG CTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCTGGTG TGAGGCAAGGG CTCAGGCTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGACAAGGGGCTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCAGGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCAGGCTGACCTCTGTCCGCGTGGGAGGGGCCGGGG TGAGGCAAGGG CTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCGGGCTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCGGGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCGGGCTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGG CTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGACAAGGGGCTCACGCTGACCTCTGTCCACGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACACTGACCTCTCTCAGCGTGGGAGGGGCCGGTG TGAGGCAAGGGGCTCACGCTGACCTCTGTCCGCGTGGGAGGGGCCGGTG TGAGGCAAGGG CTCACACTGACCTCTCTCAGCGTGGGAGGAGCCAGTG TGAGGCAGGGG CTCACGC

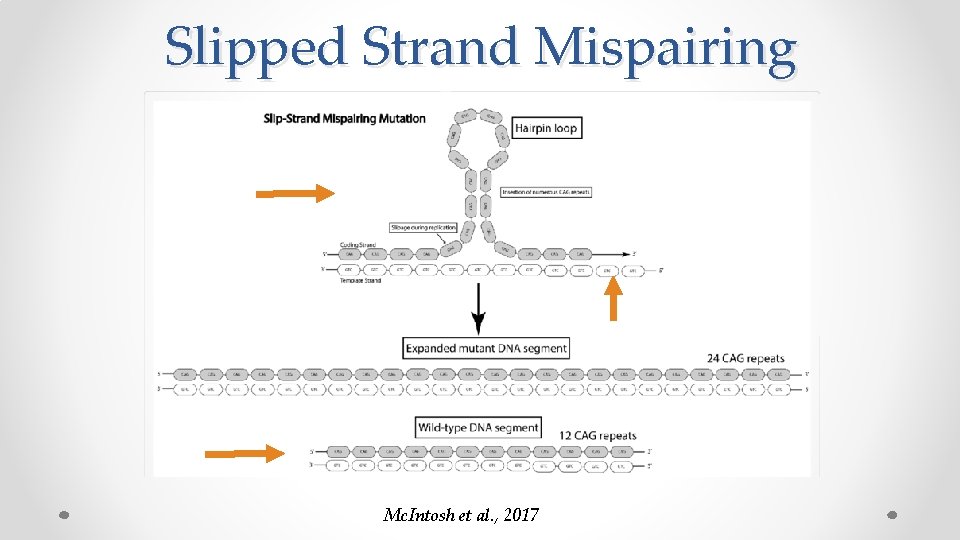
Slipped Strand Mispairing Mc. Intosh et al. , 2017

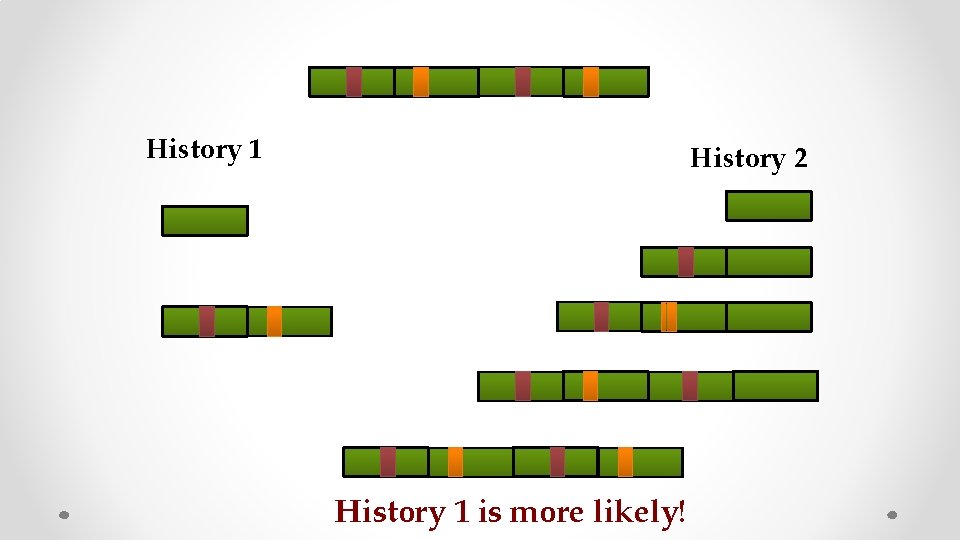
History 1 History 2 History 1 is more likely!

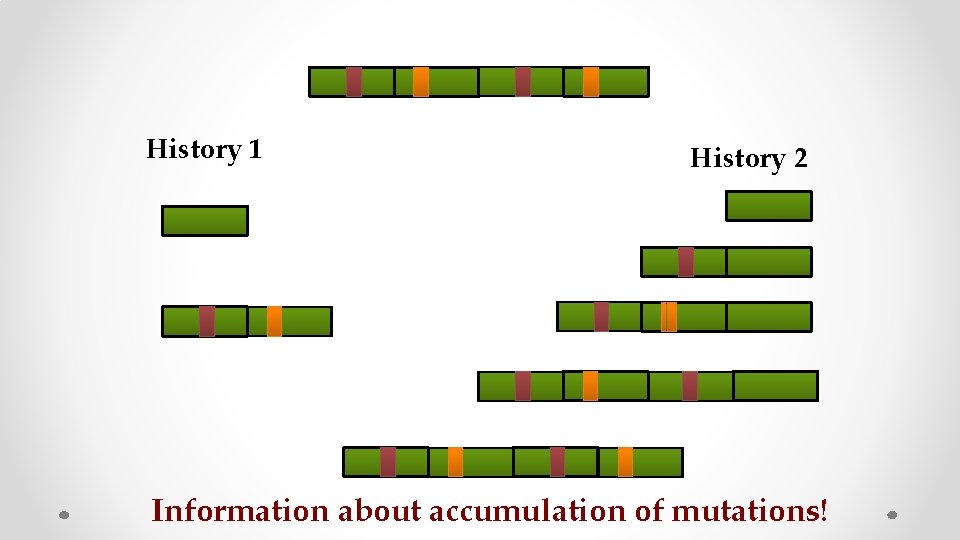
History 1 History 2 Information about accumulation of mutations!

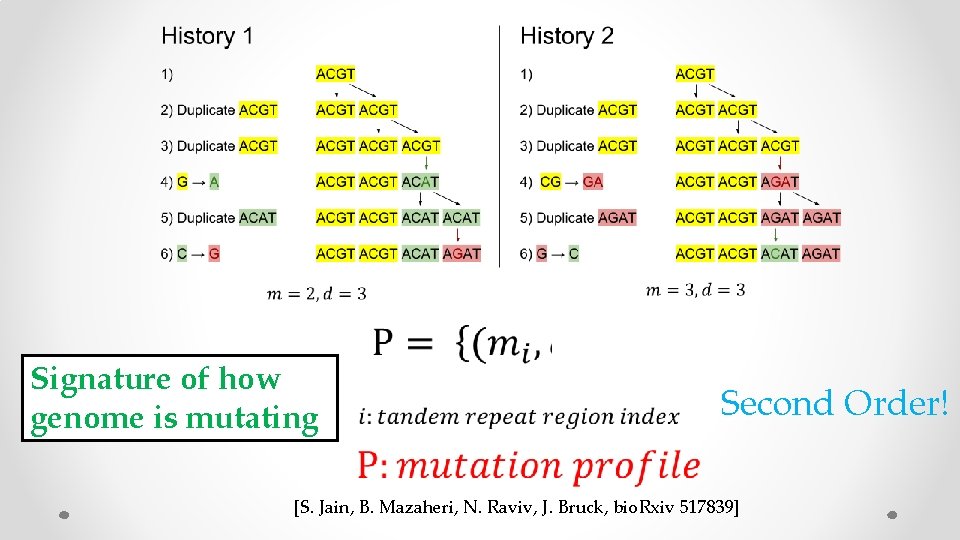
Signature of how genome is mutating Second Order! [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]

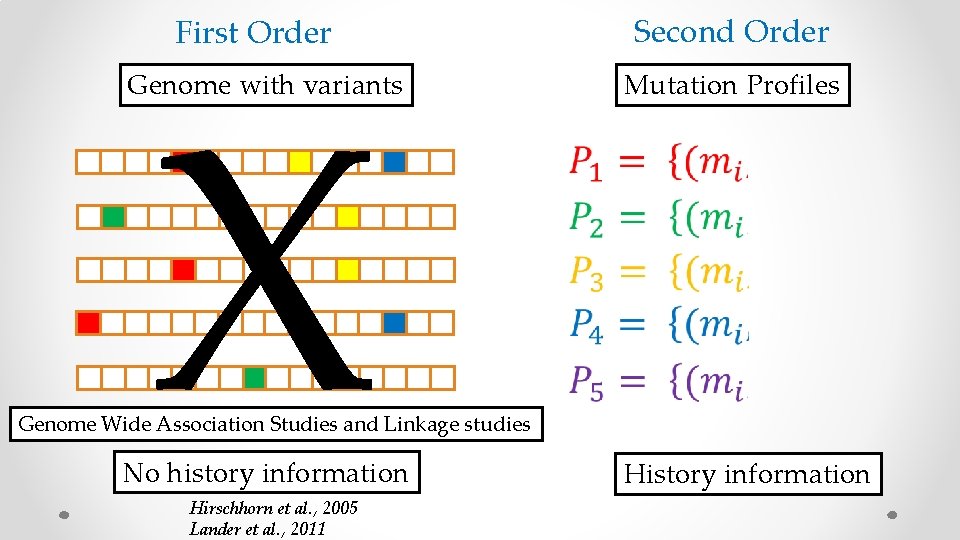
Second Order First Order X Genome with variants Mutation Profiles Genome Wide Association Studies and Linkage studies No history information Hirschhorn et al. , 2005 Lander et al. , 2011 History information

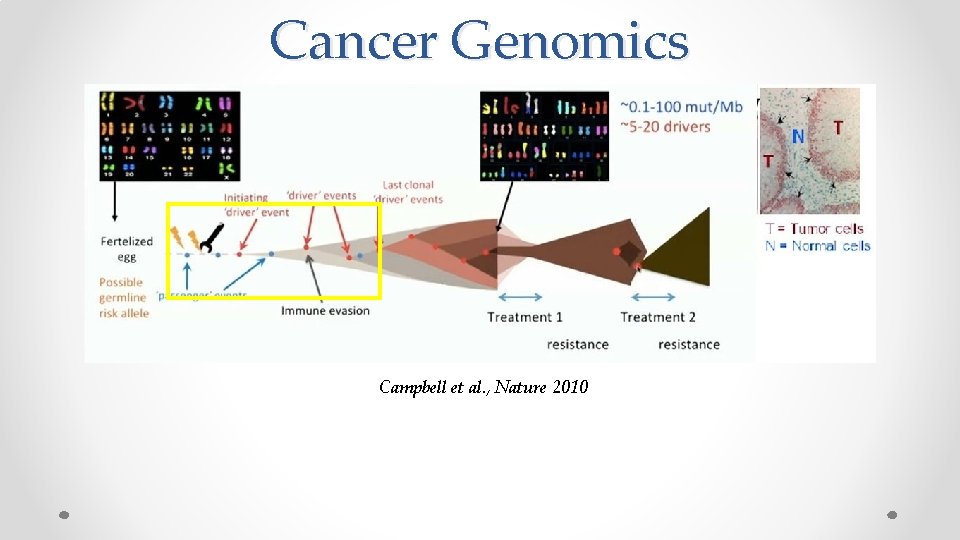
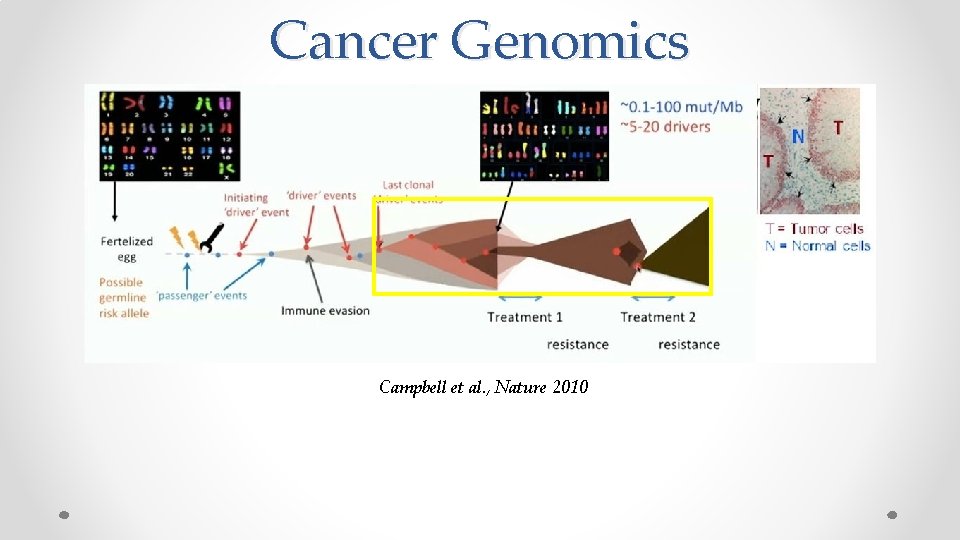
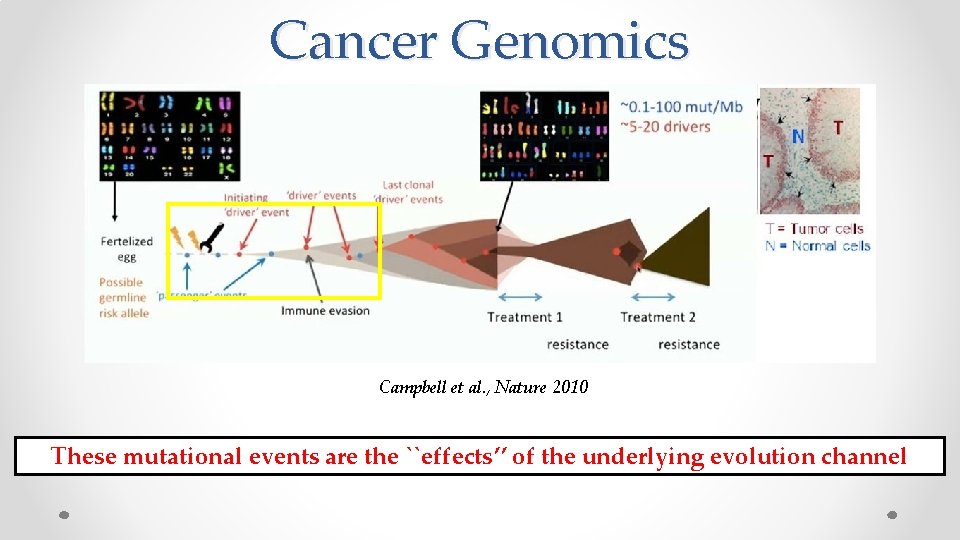
Cancer Genomics Campbell et al. , Nature 2010

Cancer Genomics Campbell et al. , Nature 2010

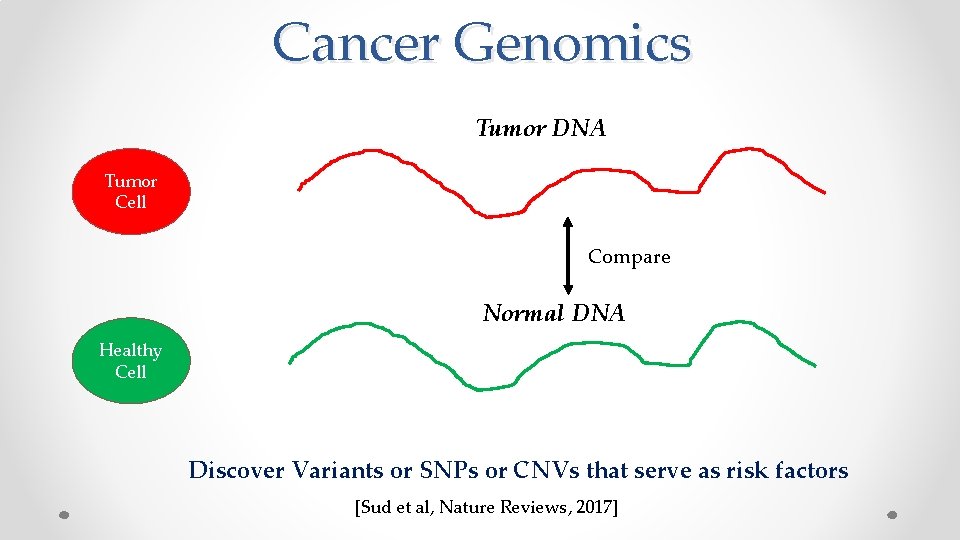
Cancer Genomics Tumor DNA Tumor Cell Compare Normal DNA Healthy Cell Discover Variants or SNPs or CNVs that serve as risk factors [Sud et al, Nature Reviews, 2017]

Cancer Genomics Campbell et al. , Nature 2010 These mutational events are the ``effects’’ of the underlying evolution channel

Hypothesis Intrinsic mutation rate associated with the propensity to accumulate driver (bad) mutations that lead to certain kind of cancer. May be the mutation profiles of normal (healthy) DNA contain some signal about this accumulation. If true, we can use mutation profiles of normal DNA to predict future cancer risk To verify the hypothesis: Need DNA of people before they got cancer. Availing such data is currently not possible.

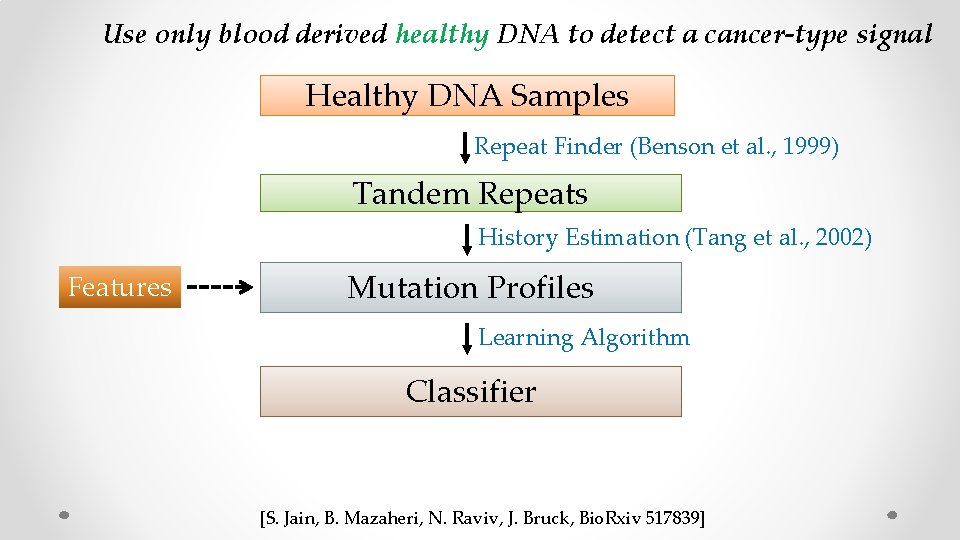
Approximation Will use blood-derived “healthy” DNA of cancer patients to check if the mutation profiles show any association with the cancer-type.

Source of Data

Use only blood derived healthy DNA to detect a cancer-type signal Healthy DNA Samples Repeat Finder (Benson et al. , 1999) Tandem Repeats History Estimation (Tang et al. , 2002) Features Mutation Profiles Learning Algorithm Classifier [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, Bio. Rxiv 517839]

• Each DNA sample file: 15 -25 GB • Number of samples: > 5000 Data! • Total data ~ 75 TB – 125 TB • Data security protocol: can only use a secure cluster • Become an ENGINEER! • Use a cluster. To learn! • Figure out the TCGA database. • Automate downloading of data. • Shell, python and C++ scripting • Learn Bioinformatics tools • Store processed data • Do some data science!

Use only blood derived healthy DNA to detect a cancer-type signal Healthy DNA Samples Repeat Finder (Benson et al. , 1999) Tandem Repeats History Estimation (Tang et al. , 2002) Features Mutation Profiles Learning Algorithm Classifier [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, Bio. Rxiv 517839]

75% training set 4 -fold cross-validation 25% test set

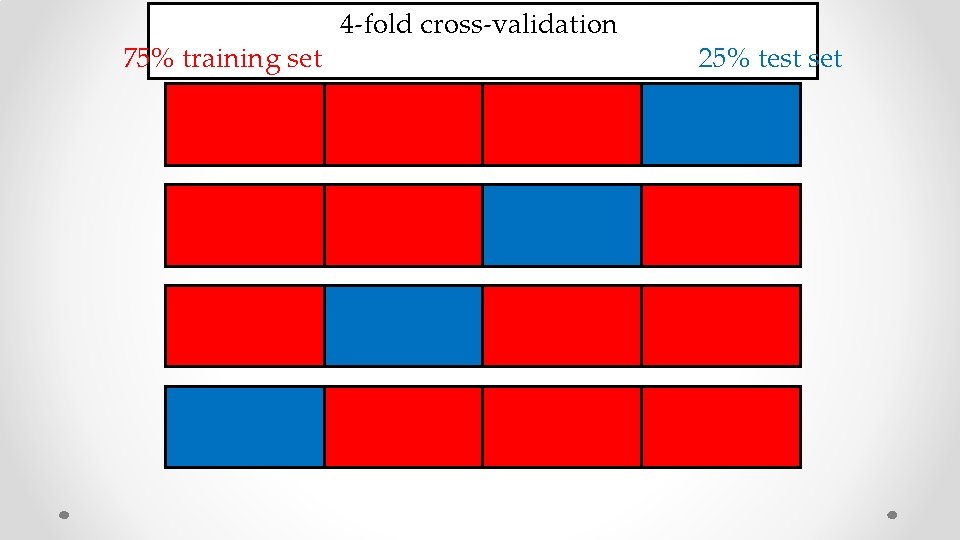
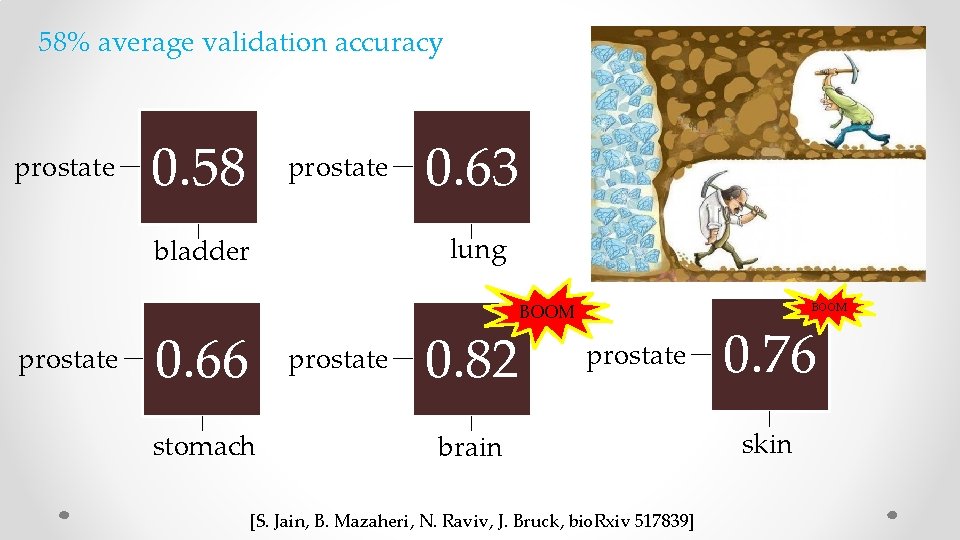
58% average validation accuracy prostate 0. 58 prostate lung bladder prostate 0. 63 BOOM 0. 66 prostate stomach 0. 82 BOOM prostate brain [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839] 0. 76 skin

Question for the audience What machine learning algorithm worked best for us to obtain the results shown ? (a) Neural Networks (b) SVM (c) Decision Tree – Gradient Boosting

Question for the audience What machine learning algorithm worked best for us to obtain the results shown ? Decision Tree – Gradient Boosting Brieman et al. , 1984 Mason et al. , 1999
![S Jain B Mazaheri N Raviv J Bruck bio Rxiv 517839 [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-92.jpg)
[S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]
![S Jain B Mazaheri N Raviv J Bruck bio Rxiv 517839 [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-93.jpg)
[S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]
![Class1 brain Class2 skin Class3 pancreas Class4 rest S Jain Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Class_4 = [rest] [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-94.jpg)
Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Class_4 = [rest] [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]
![Class1 brain Class2 skin Class3 pancreas Healthy Genome Multiclassifier S Jain Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-95.jpg)
Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]
![Class1 brain Class2 skin Class3 pancreas Healthy Genome Multiclassifier S Jain Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-96.jpg)
Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]
![Class1 brain Class2 skin Class3 pancreas Healthy Genome Multiclassifier S Jain Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain,](https://slidetodoc.com/presentation_image/58d83e78872b1bbec5e06eba9c948257/image-97.jpg)
Class_1 = [brain] Class_2 = [skin] Class_3 = [pancreas] Healthy Genome Multiclassifier [S. Jain, B. Mazaheri, N. Raviv, J. Bruck, bio. Rxiv 517839]

Summary • New microscope to view the genome. • Decodes the evolutionary memory of tandem repeat regions to measure the accumulation of mutations. • Detected the cancer-type signal from the healthy genome. • Implicitly inferring about a process of acquiring mutations in the blood that is associated with cancer in a tissue-specific way. • Has potential applications in predicting future cancer risk and early cancer detection. https: //www. biorxiv. org/content/10. 1101/517839 v 1

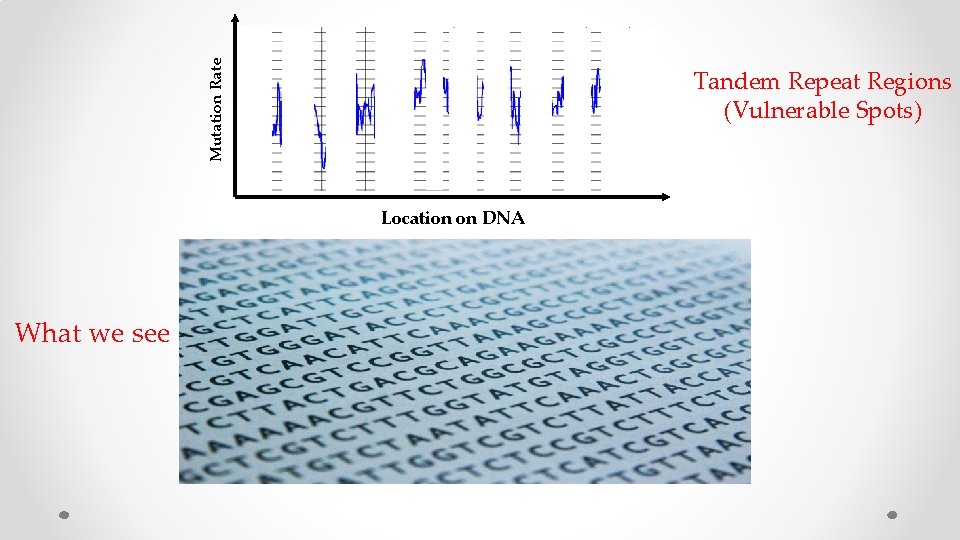
Mutation Rate MASK Location on DNA What we see

Mutation Rate Tandem Repeat Regions (Vulnerable Spots) Location on DNA What we see

Thank you Announcements!

Programming Challenge Thursday, 05/30, 2. 30 pm – PCP 1. Joseph Min 2. Ajay Natarajan & Ananth Malladi & Shubh Agarwal 3. Monika Getsova 4. Justin Zhang & Sebastien Abadi 5. Kade Imanaka & Selina Zhou

Everyone has a Gift! Tuesday, 06/04, 2. 30 pm – MQ 1 1. Toussaint Pegues 2. Paromita Mitchell 3. Maya Joysula 4. Thomas Barrett 5. Polina Verkhovodova 6. Jeffrey Ma 7. Kade Immanka

2000 2020 2040 Thursday, 06/06, 2. 30 pm – MQ 2 1. Tatiana Brailovskaya 2. Isabella Camplisson & Chan Gi Kim 3. Colin Chun 4. Michelle M Hyun & Isaac John Perrin 5. Madison Lee 6. Vincent Tieu 7. Nora Griffith 8. Ananth Malladi & Forrest Graham