Evolution of protein coding sequences Kinds of nucleotide




- Slides: 32

Evolution of protein coding sequences

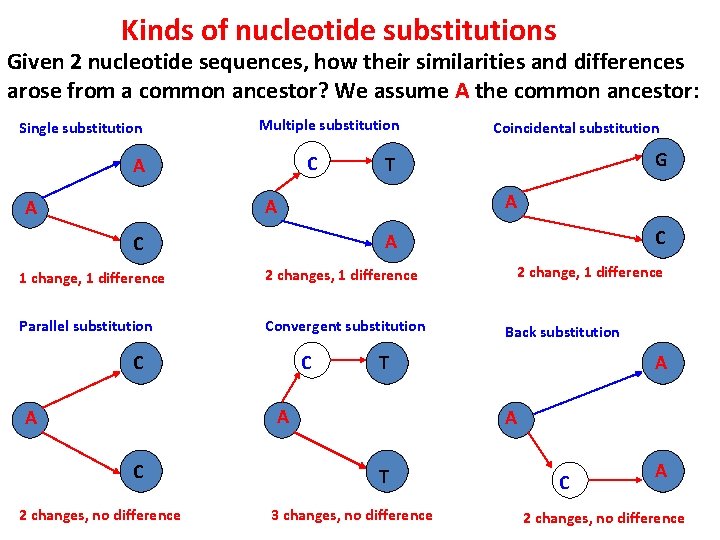
Kinds of nucleotide substitutions Given 2 nucleotide sequences, how their similarities and differences arose from a common ancestor? We assume A the common ancestor: Single substitution Multiple substitution C A A 1 change, 1 difference 2 changes, 1 difference Parallel substitution Convergent substitution C C 2 changes, no difference 2 change, 1 difference Back substitution T A C C A G T A A Coincidental substitution A A T 3 changes, no difference C A 2 changes, no difference

Important properties inherent to the standard genetic code

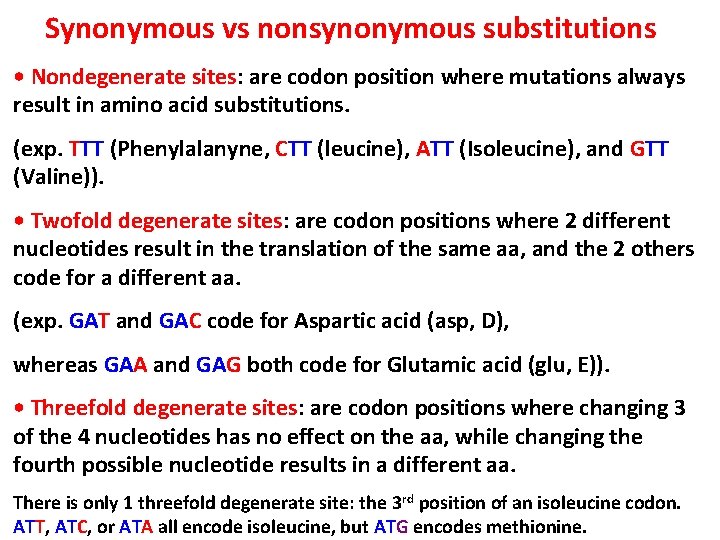
Synonymous vs nonsynonymous substitutions • Nondegenerate sites: are codon position where mutations always result in amino acid substitutions. (exp. TTT (Phenylalanyne, CTT (leucine), ATT (Isoleucine), and GTT (Valine)). • Twofold degenerate sites: are codon positions where 2 different nucleotides result in the translation of the same aa, and the 2 others code for a different aa. (exp. GAT and GAC code for Aspartic acid (asp, D), whereas GAA and GAG both code for Glutamic acid (glu, E)). • Threefold degenerate sites: are codon positions where changing 3 of the 4 nucleotides has no effect on the aa, while changing the fourth possible nucleotide results in a different aa. There is only 1 threefold degenerate site: the 3 rd position of an isoleucine codon. ATT, ATC, or ATA all encode isoleucine, but ATG encodes methionine.

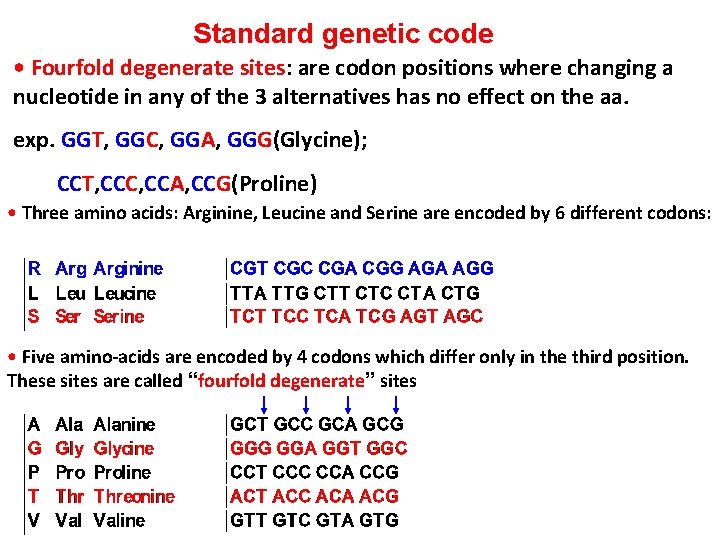
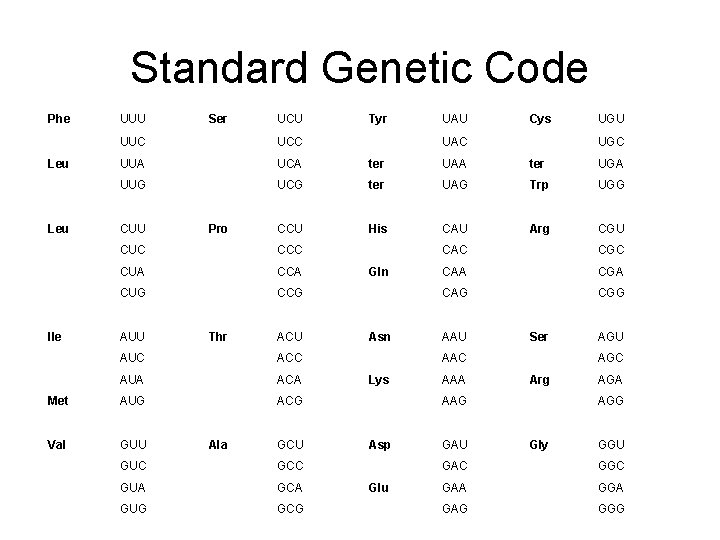
Standard genetic code • Fourfold degenerate sites: are codon positions where changing a nucleotide in any of the 3 alternatives has no effect on the aa. exp. GGT, GGC, GGA, GGG(Glycine); CCT, CCC, CCA, CCG(Proline) • Three amino acids: Arginine, Leucine and Serine are encoded by 6 different codons: • Five amino-acids are encoded by 4 codons which differ only in the third position. These sites are called “fourfold degenerate” sites

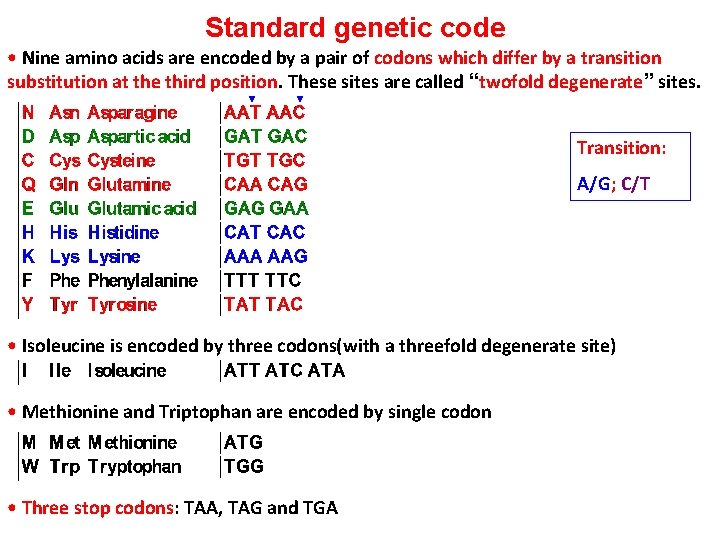
Standard genetic code • Nine amino acids are encoded by a pair of codons which differ by a transition substitution at the third position. These sites are called “twofold degenerate” sites. Transition: A/G; C/T • Isoleucine is encoded by three codons(with a threefold degenerate site) • Methionine and Triptophan are encoded by single codon • Three stop codons: TAA, TAG and TGA

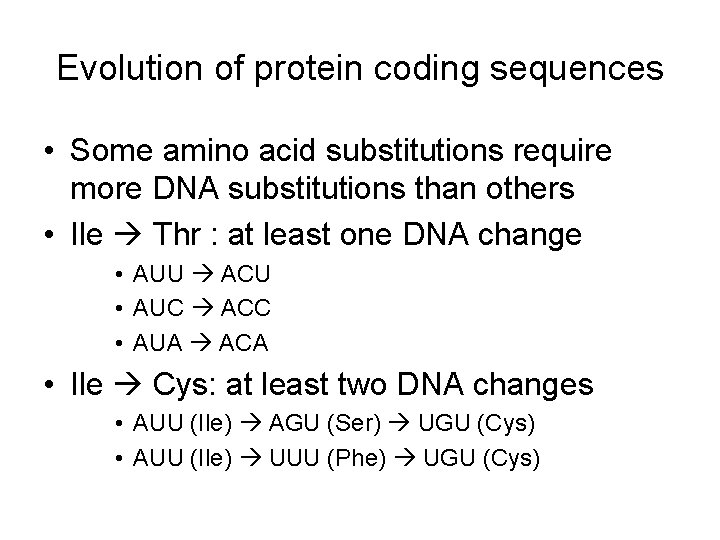
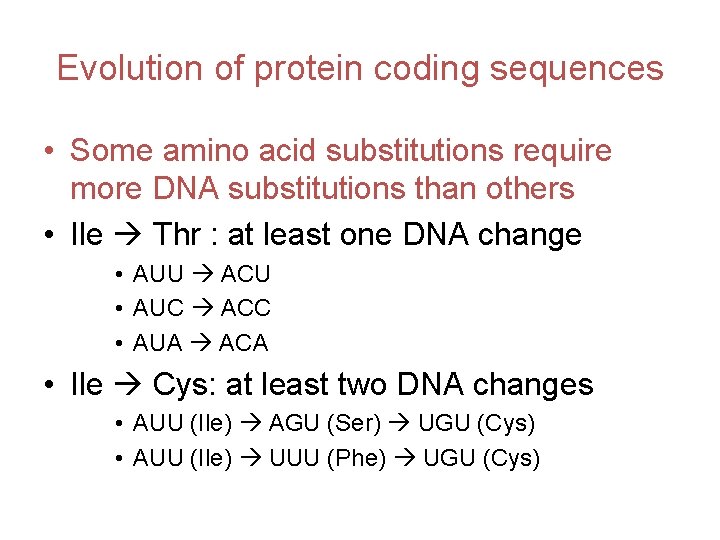
Evolution of protein coding sequences • Some amino acid substitutions require more DNA substitutions than others • Ile Thr : at least one DNA change • AUU ACU • AUC ACC • AUA ACA • Ile Cys: at least two DNA changes • AUU (Ile) AGU (Ser) UGU (Cys) • AUU (Ile) UUU (Phe) UGU (Cys)

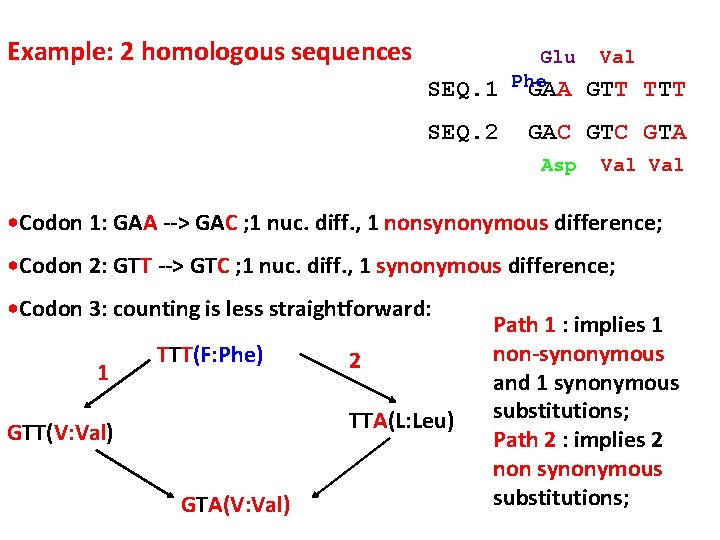
Example: 2 homologous sequences SEQ. 1 SEQ. 2 Glu Phe GAA Val GTT TTT GAC GTA Asp Val • Codon 1: GAA --> GAC ; 1 nuc. diff. , 1 nonsynonymous difference; • Codon 2: GTT --> GTC ; 1 nuc. diff. , 1 synonymous difference; • Codon 3: counting is less straightforward: 1 TTT(F: Phe) 2 TTA(L: Leu) GTT(V: Val) GTA(V: Val) Path 1 : implies 1 non-synonymous and 1 synonymous substitutions; Path 2 : implies 2 non synonymous substitutions;

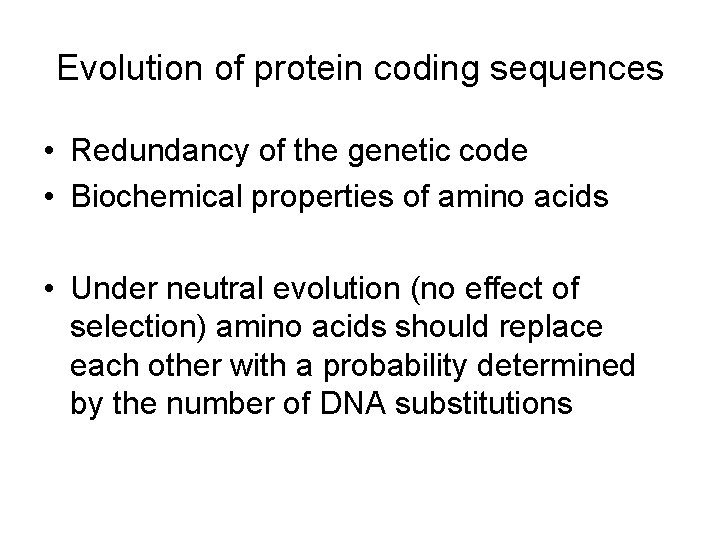
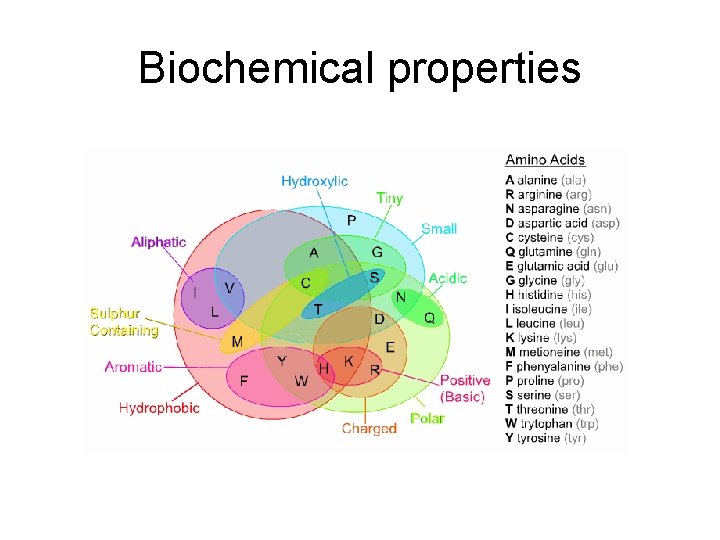
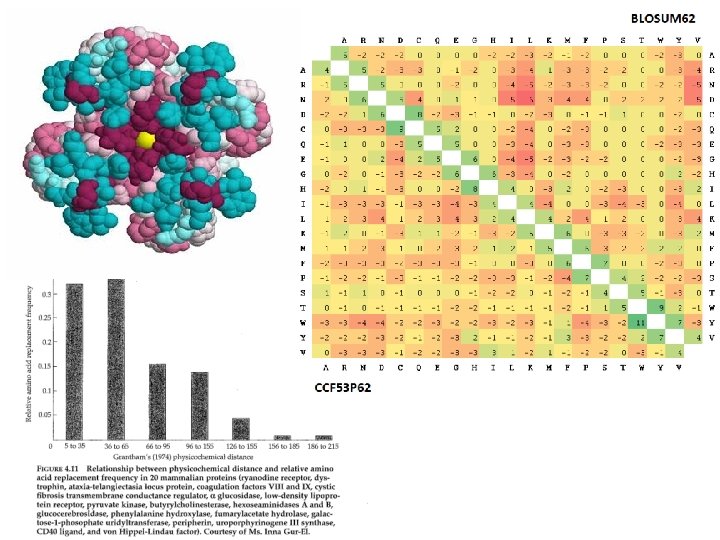
Evolution of protein coding sequences • Redundancy of the genetic code • Biochemical properties of amino acids • Under neutral evolution (no effect of selection) amino acids should replace each other with a probability determined by the number of DNA substitutions

Evolution of protein coding sequences • Some amino acid substitutions require more DNA substitutions than others • Ile Thr : at least one DNA change • AUU ACU • AUC ACC • AUA ACA • Ile Cys: at least two DNA changes • AUU (Ile) AGU (Ser) UGU (Cys) • AUU (Ile) UUU (Phe) UGU (Cys)

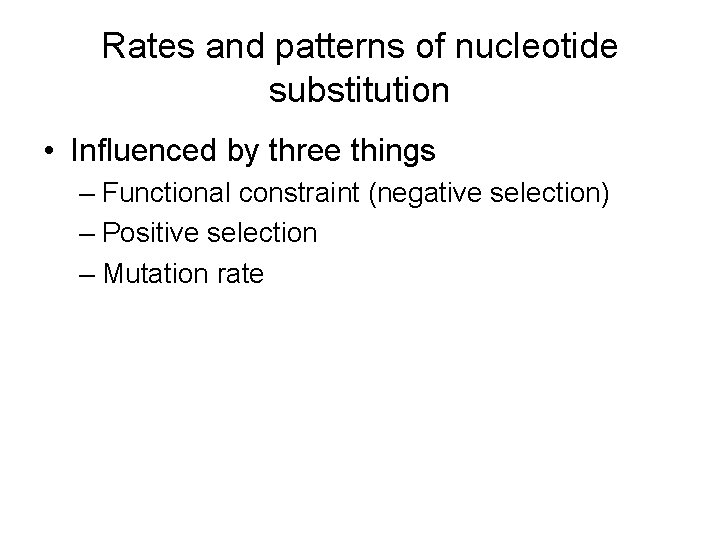
Rates and patterns of nucleotide substitution • Influenced by three things – Functional constraint (negative selection) – Positive selection – Mutation rate

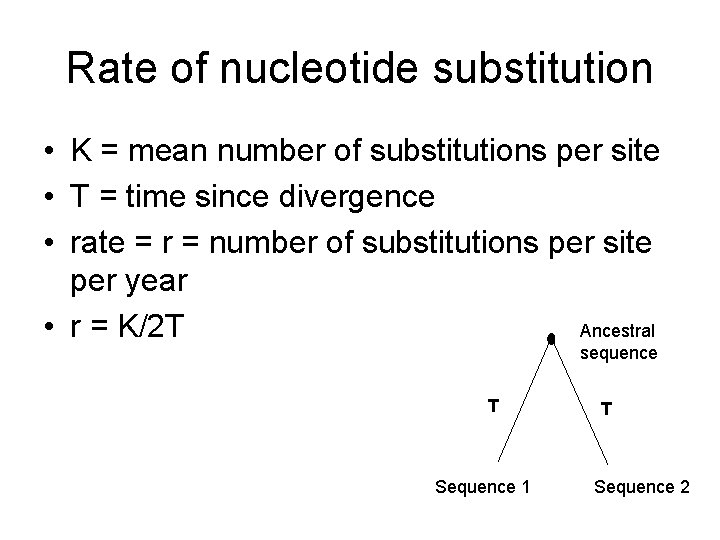
Rate of nucleotide substitution • K = mean number of substitutions per site • T = time since divergence • rate = r = number of substitutions per site per year Ancestral • r = K/2 T sequence T Sequence 1 T Sequence 2

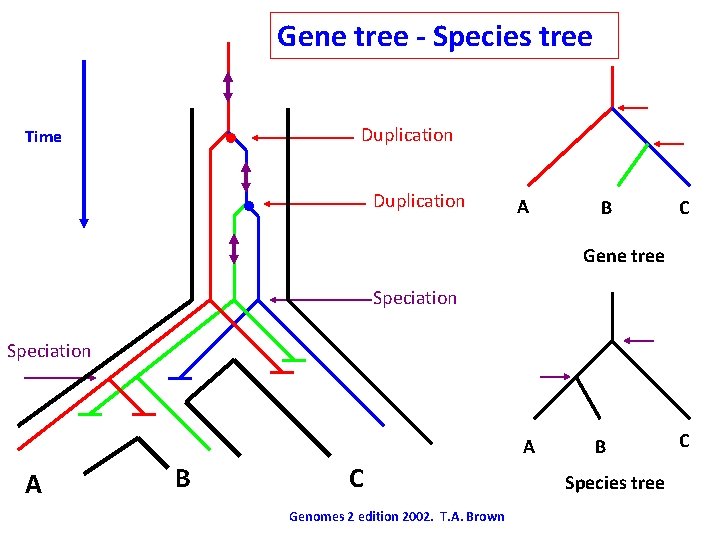
Gene tree - Species tree • Time Duplication • Duplication A B C Gene tree Speciation A A B C Genomes 2 edition 2002. T. A. Brown B Species tree C

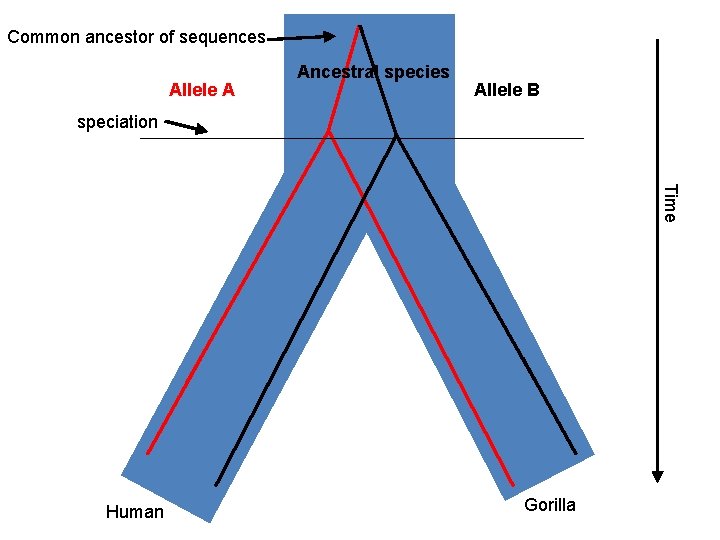
Common ancestor of sequences Allele A Ancestral species Allele B speciation Time Human Gorilla

Evolution of protein-coding sequences • The Genetic Code is redundant • Some nucleotide changes do not change the amino acid coded for – 3 rd codon position often synonymous – 2 nd position never – 1 st position sometimes

Standard Genetic Code Phe Leu Ile UUU Ser UCU Tyr UAU Cys UUC UCC UUA UCA ter UAA ter UGA UUG UCG ter UAG Trp UGG CCU His CAU Arg CGU CUU Pro CUC CCC CUA CCA CUG CCG AUU Thr ACU AUC ACC AUA ACA Met AUG ACG Val GUU Ala GCU GUC GCC GUA GCA GUG GCG UAC UGU Gln Asn UGC CAC CGC CAA CGA CAG CGG AAU Ser AAC Lys AAA AGC Arg AAG Asp Glu GAU AGA AGG Gly GGU GAC GGC GAA GGA GAG GGG

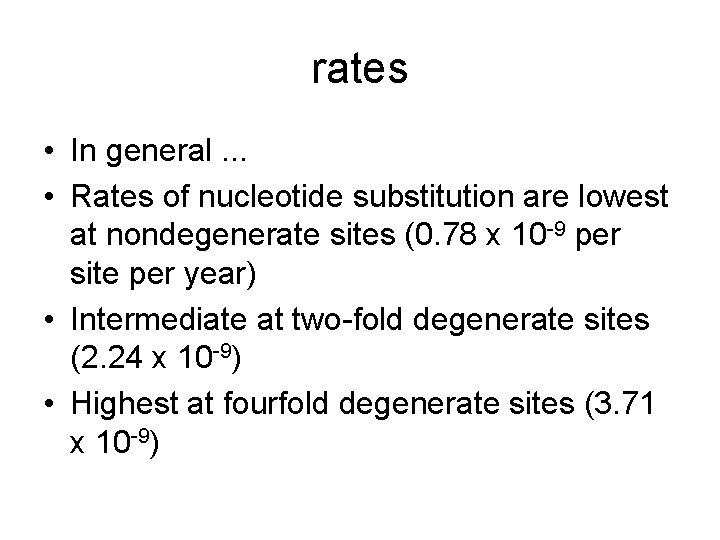
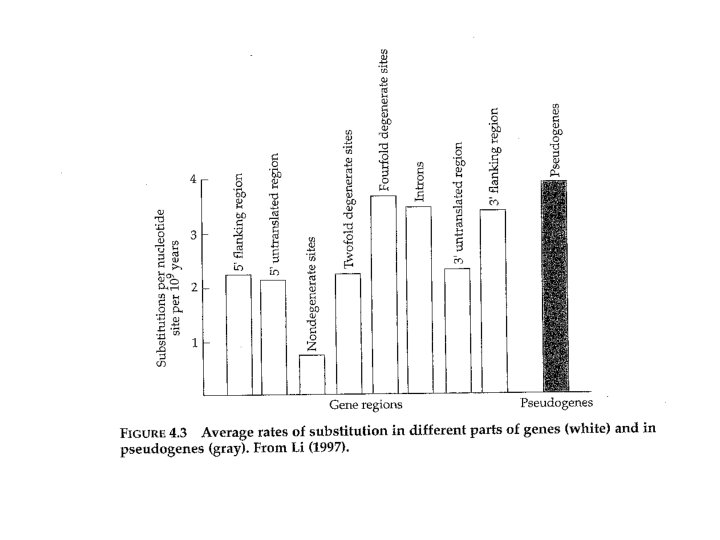
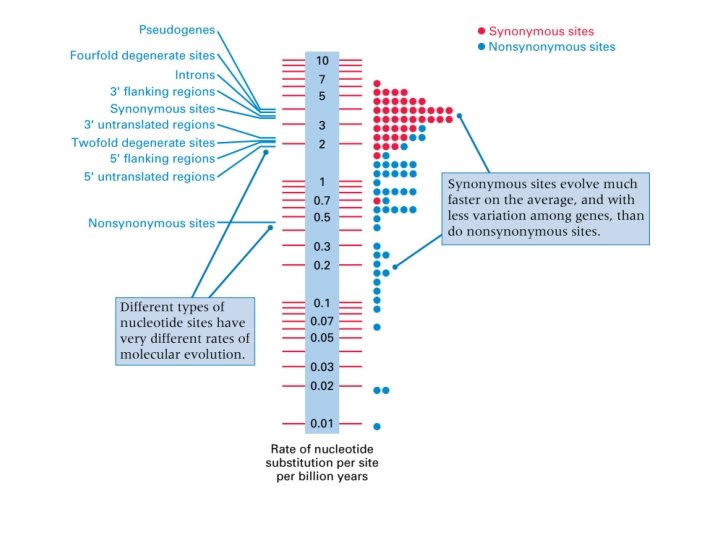
rates • In general. . . • Rates of nucleotide substitution are lowest at nondegenerate sites (0. 78 x 10 -9 per site per year) • Intermediate at two-fold degenerate sites (2. 24 x 10 -9) • Highest at fourfold degenerate sites (3. 71 x 10 -9)

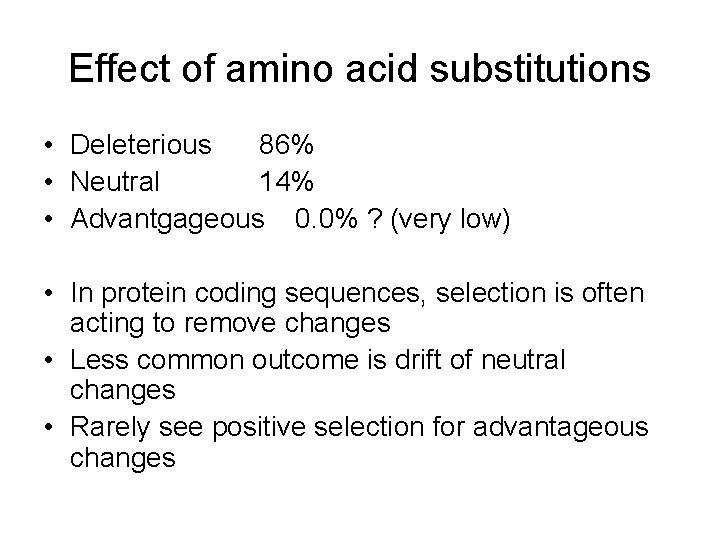
Effect of amino acid substitutions • Deleterious 86% • Neutral 14% • Advantgageous 0. 0% ? (very low) • In protein coding sequences, selection is often acting to remove changes • Less common outcome is drift of neutral changes • Rarely see positive selection for advantageous changes

Functional Constraint • Proteins often have some functional constraint • The stronger the functional constraint, the slower the rate of evolution

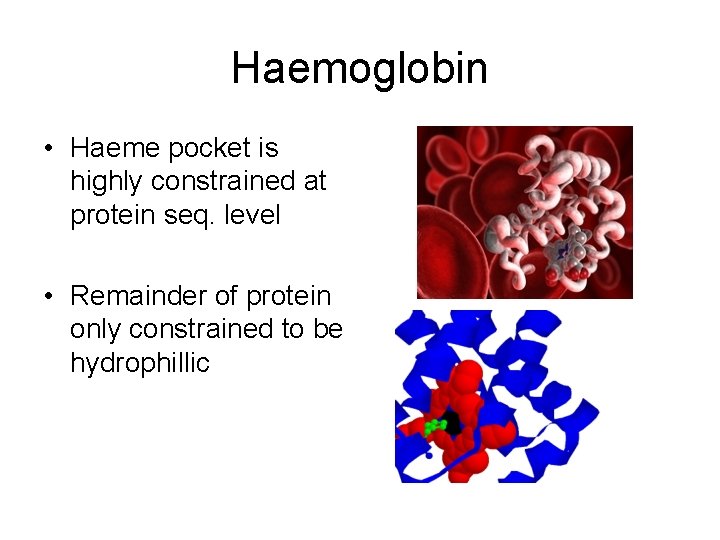
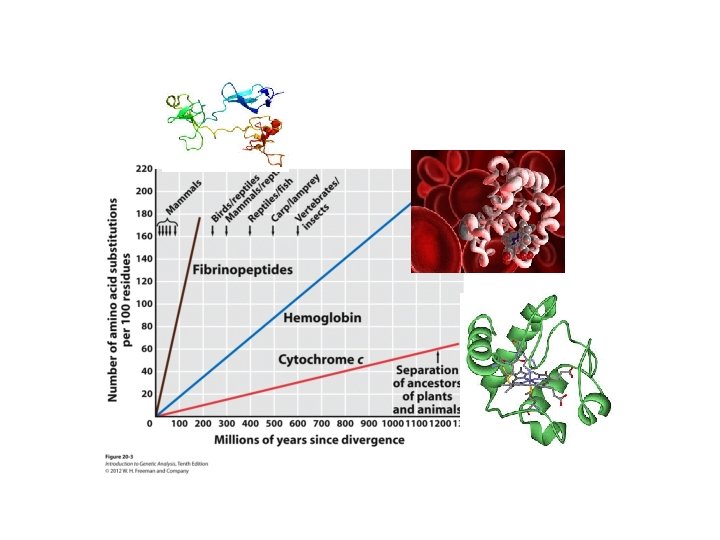
Haemoglobin • Haeme pocket is highly constrained at protein seq. level • Remainder of protein only constrained to be hydrophillic

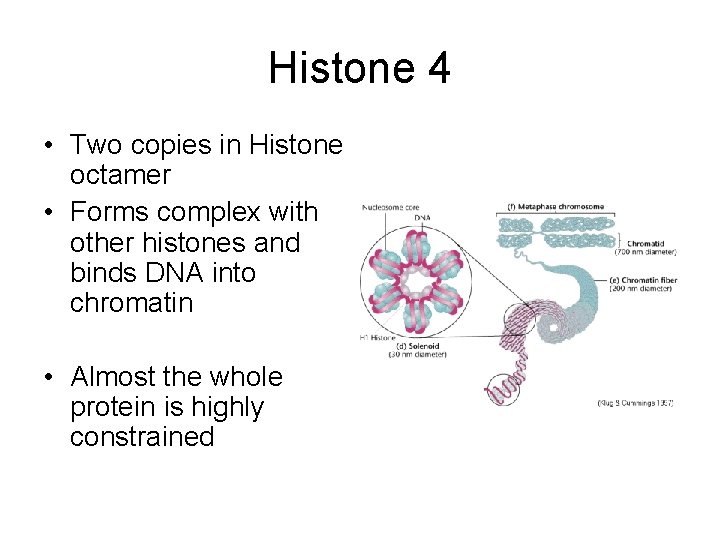
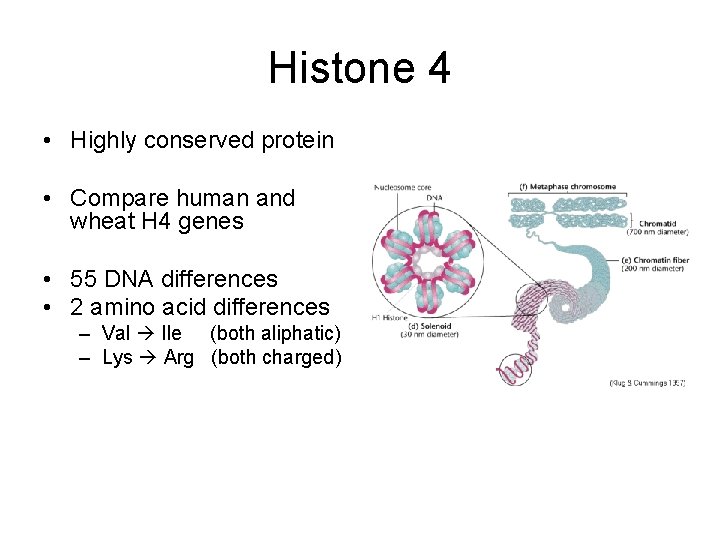
Histone 4 • Two copies in Histone octamer • Forms complex with other histones and binds DNA into chromatin • Almost the whole protein is highly constrained

Fibrinopeptides • Hardly any sequence constraint


Rates and Patterns • Patterns of change can be informative of the function of a protein • Different genes evolve at different rates • Amino acids that are always conserved are likely to be critical to the function

Biochemical properties


Histone 4 • Highly conserved protein • Compare human and wheat H 4 genes • 55 DNA differences • 2 amino acid differences – Val Ile (both aliphatic) – Lys Arg (both charged)

Evolution of non-coding regions • homologous sequences • e. g. , compare introns of homologous genes • 5’ UTR and 3’ UTR (untranslated region) • Pseudogenes



Synonymous substitution rate variation • Synonymous rates may differ between genes • How come? • Maybe different mutation rates in different parts of the genome

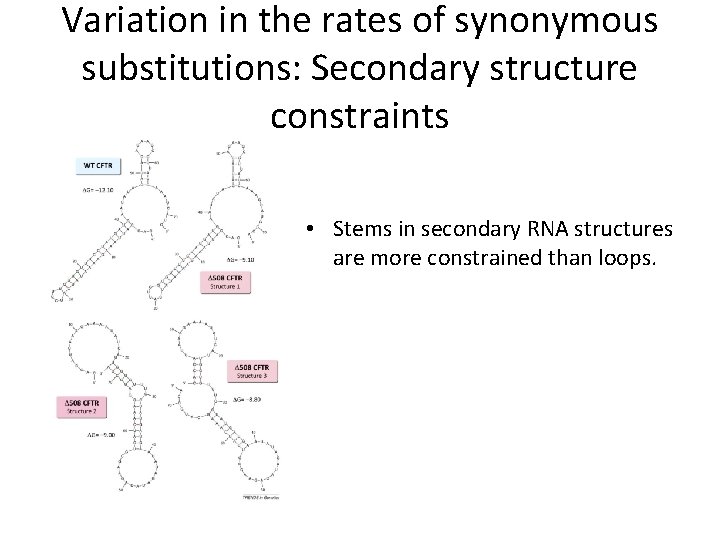
Variation in the rates of synonymous substitutions: Secondary structure constraints • Stems in secondary RNA structures are more constrained than loops.