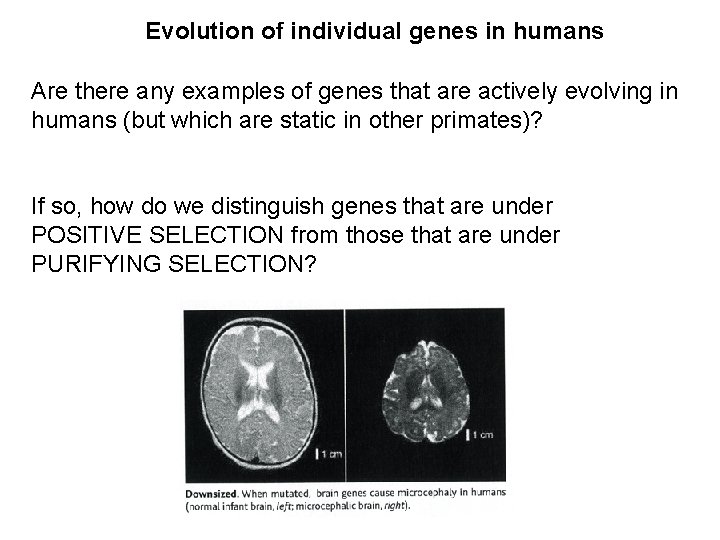
Evolution of individual genes in humans Are there



- Slides: 14

Evolution of individual genes in humans Are there any examples of genes that are actively evolving in humans (but which are static in other primates)? If so, how do we distinguish genes that are under POSITIVE SELECTION from those that are under PURIFYING SELECTION?

Four papers to read: 1. 2. Balter, M. 2005. Are human brains still evolving? SCIENCE 309: 1662 - 1663. 2. Evans, P. D. et al. 2004. Reconstructing the evolutionary history of microcephalin, a gene controlling human brain size. Human Molecular Genetics 13: 1139 -1145. 3. 4. Vallender, E. J. and B. T. Lahn. 2004. Positive selection on the human genome. Human Molecular Genetics 13: R 245 -R 254. Evans, P. D. et al. 2005. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. SCIENCE 309: 1717 -1720.

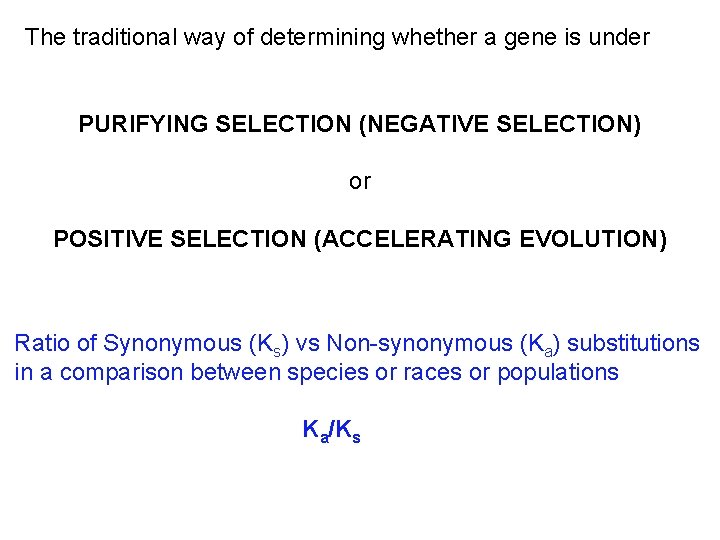
The traditional way of determining whether a gene is under PURIFYING SELECTION (NEGATIVE SELECTION) or POSITIVE SELECTION (ACCELERATING EVOLUTION) Ratio of Synonymous (Ks) vs Non-synonymous (Ka) substitutions in a comparison between species or races or populations Ka/Ks

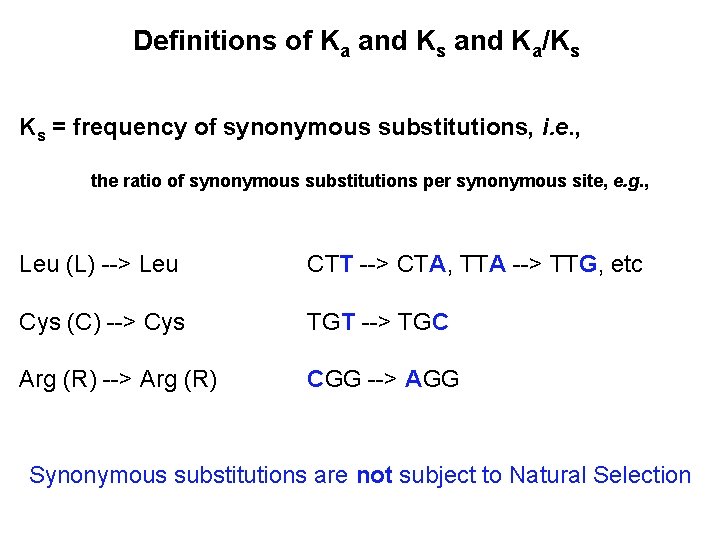
Definitions of Ka and Ks and Ka/Ks Ks = frequency of synonymous substitutions, i. e. , the ratio of synonymous substitutions per synonymous site, e. g. , Leu (L) --> Leu CTT --> CTA, TTA --> TTG, etc Cys (C) --> Cys TGT --> TGC Arg (R) --> Arg (R) CGG --> AGG Synonymous substitutions are not subject to Natural Selection

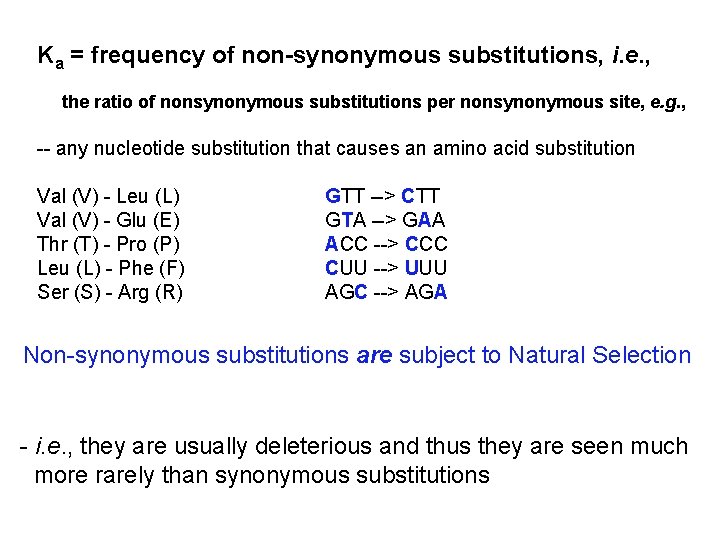
Ka = frequency of non-synonymous substitutions, i. e. , the ratio of nonsynonymous substitutions per nonsynonymous site, e. g. , -- any nucleotide substitution that causes an amino acid substitution Val (V) - Leu (L) Val (V) - Glu (E) Thr (T) - Pro (P) Leu (L) - Phe (F) Ser (S) - Arg (R) GTT --> CTT GTA --> GAA ACC --> CCC CUU --> UUU AGC --> AGA Non-synonymous substitutions are subject to Natural Selection - i. e. , they are usually deleterious and thus they are seen much more rarely than synonymous substitutions

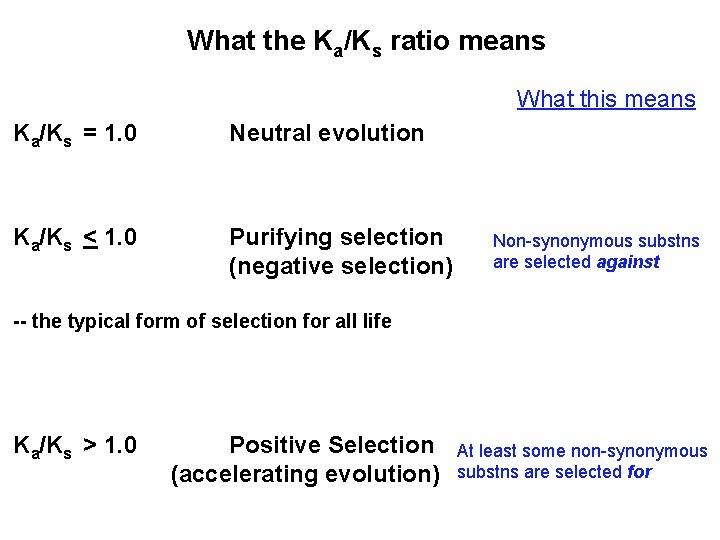
What the Ka/Ks ratio means What this means Ka/Ks = 1. 0 Neutral evolution Ka/Ks < 1. 0 Purifying selection (negative selection) Non-synonymous substns are selected against -- the typical form of selection for all life Ka/Ks > 1. 0 Positive Selection (accelerating evolution) At least some non-synonymous substns are selected for

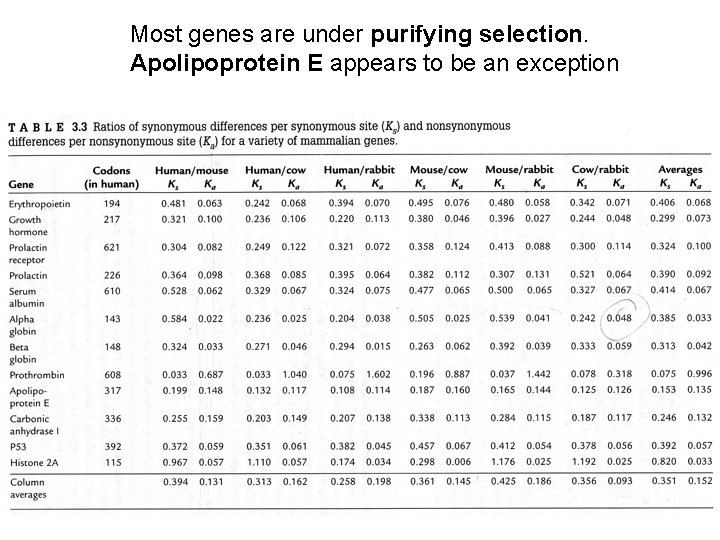
Most genes are under purifying selection. Apolipoprotein E appears to be an exception

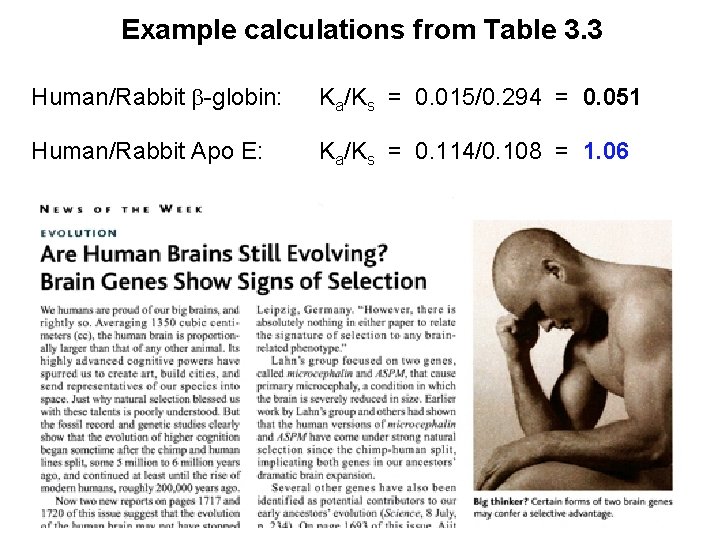
Example calculations from Table 3. 3 Human/Rabbit -globin: Ka/Ks = 0. 015/0. 294 = 0. 051 Human/Rabbit Apo E: Ka/Ks = 0. 114/0. 108 = 1. 06


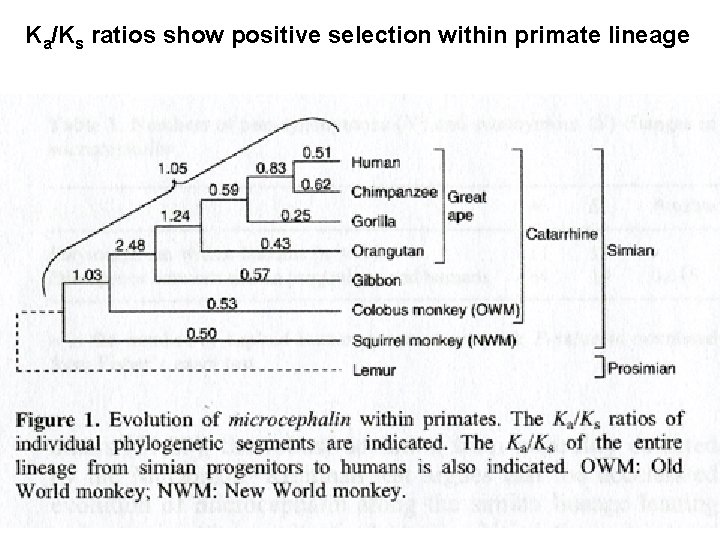
Ka/Ks ratios show positive selection within primate lineage

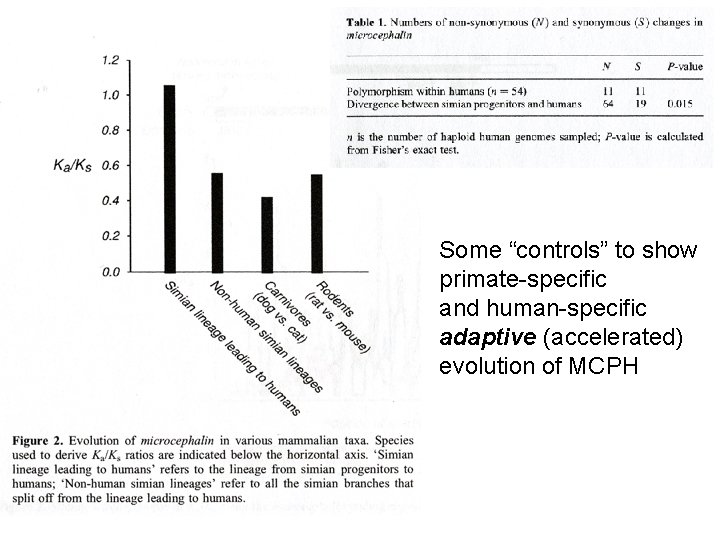
Some “controls” to show primate-specific and human-specific adaptive (accelerated) evolution of MCPH

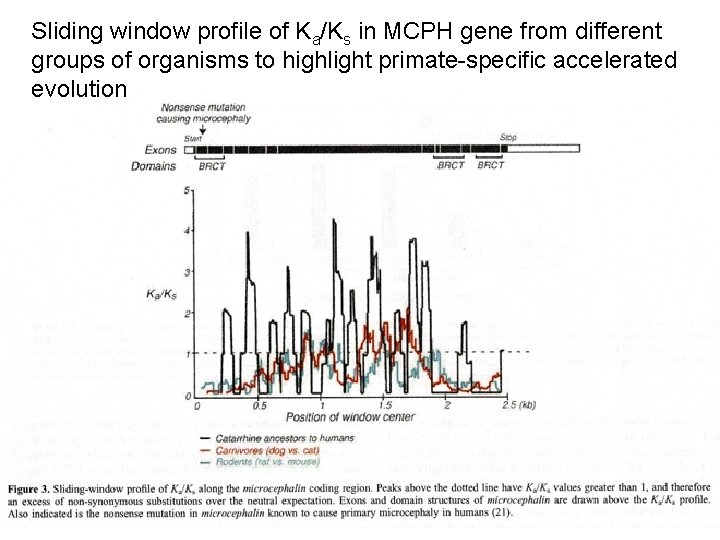
Sliding window profile of Ka/Ks in MCPH gene from different groups of organisms to highlight primate-specific accelerated evolution

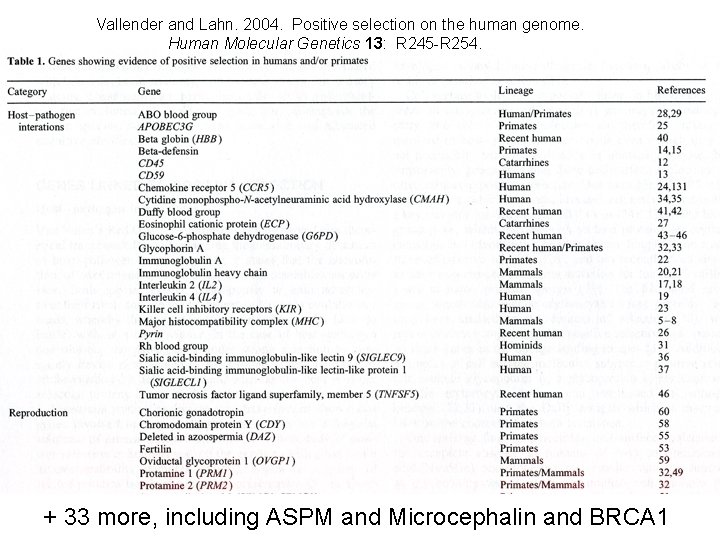
Vallender and Lahn. 2004. Positive selection on the human genome. Human Molecular Genetics 13: R 245 -R 254. + 33 more, including ASPM and Microcephalin and BRCA 1
