ESI and MALDI LCMSMS Approaches for Larger Scale


- Slides: 17

ESI and MALDI LC/MS-MS Approaches for Larger Scale Protein Identification and Quantification: Are They Equivalent? 1 P. Juhasz, 1 A. Falick, 1 A. Graber, 1 S. Hattan, 1 N. Khainovski, 1 J. Marchese, 1 S. Martin, 1 D. Patterson, 1 B. Williamson, 2 J. Malmstrom, 2 G. Westergren-Thorsson, 2 G. Marko-Varga 1 Applied Biosystems, Proteomics Research Center, Framingham, MA 2 University of Lund, Molecular Biology Dept. , Lund, Sweden

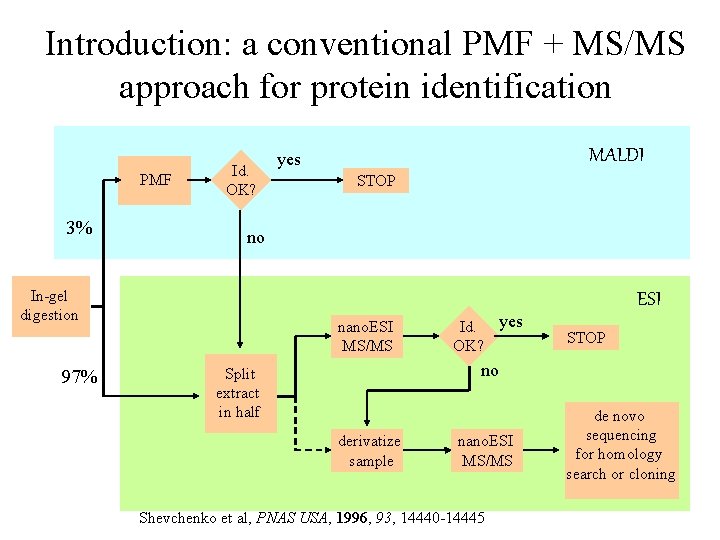
Introduction: a conventional PMF + MS/MS approach for protein identification PMF 3% Id. OK? STOP no In-gel digestion 97% MALDI yes nano. ESI MS/MS Id. yes OK? ESI STOP no Split extract in half derivatize sample nano. ESI MS/MS Shevchenko et al, PNAS USA, 1996, 93, 14440 -14445 de novo sequencing for homology search or cloning

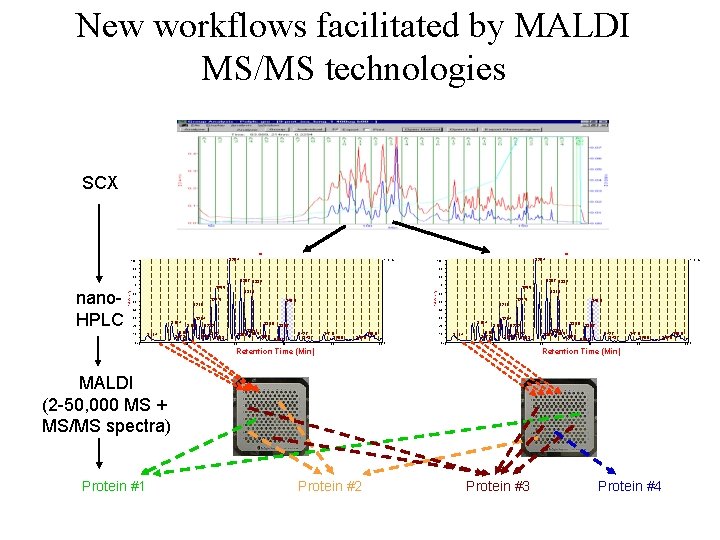
New workflows facilitated by MALDI MS/MS technologies SCX TIC T 28. 4 100 1. 7 E+4 90 T 31. 5 60 T 24. 5 50 T 30. 7 T 33. 2 70 T 25. 5 % Intensity 80 T 30. 7 T 33. 2 70 T 40. 0 T 21. 0 40 T 25. 5 T 31. 5 60 T 24. 5 50 T 40. 0 T 21. 0 40 30 T 16. 4 20 10 0 1. 7 E+4 90 80 nano. HPLC T 28. 4 100 T 11. 4 10 30 T 21. 4 T 19. 7 T 18. 1 T 17. 3 20 T 35. 6 T 23. 2 T 24. 2 T 22. 0 T 25. 3 T 32. 1 T 30. 0 T 32. 5 30 T 34. 9 T 38. 7 T 37. 7 T 16. 4 20 T 42. 7 T 43. 7 40 T 47. 9 T 49. 9 T 56. 8 10 T 54. 6 50 60 0 0 T 11. 4 10 T 21. 4 T 19. 7 T 18. 1 T 17. 3 T 35. 6 T 23. 2 T 22. 0 T 24. 2 T 25. 3 20 Retention Time (Min) T 32. 1 T 30. 0 T 32. 5 30 T 34. 9 T 38. 7 T 37. 7 T 42. 7 T 43. 7 40 T 47. 9 T 49. 9 50 Retention Time (Min) MALDI (2 -50, 000 MS + MS/MS spectra) Protein #1 Protein #2 Protein #3 Protein #4 T 56. 8 T 54. 6 60 0

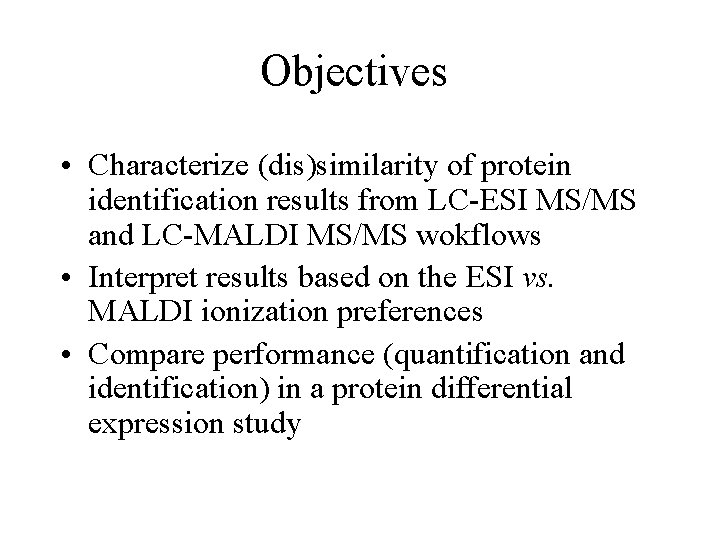
Objectives • Characterize (dis)similarity of protein identification results from LC-ESI MS/MS and LC-MALDI MS/MS wokflows • Interpret results based on the ESI vs. MALDI ionization preferences • Compare performance (quantification and identification) in a protein differential expression study

Case Study 1: Haemophilus ducreyi • H. ducreyi is a gram-negative bacterium that causes the sexually transmitted disease chancroid. • Linked to the heterosexual transmission of HIV in developing countries. • The sequencing of this genome (1. 7 Mb) has recently been completed and homology with H. influenzae is known. • A proteomic study was undertaken to help with the sequence alignment and annotation. http: //www. microbial-pathogenesis. org/H. ducreyi/

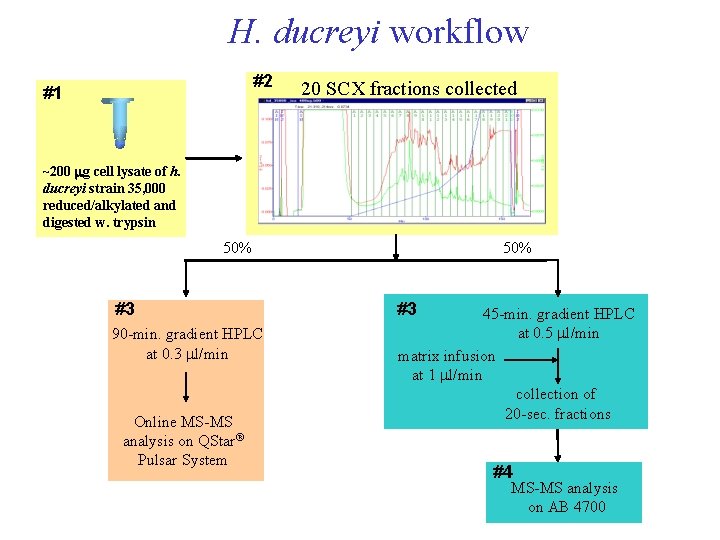
H. ducreyi workflow #2 #1 20 SCX fractions collected ~200 mg cell lysate of h. ducreyi strain 35, 000 reduced/alkylated and digested w. trypsin 50% #3 90 -min. gradient HPLC at 0. 3 ml/min Online MS-MS analysis on QStar® Pulsar System 50% #3 45 -min. gradient HPLC at 0. 5 ml/min matrix infusion at 1 ml/min collection of 20 -sec. fractions #4 MS-MS analysis on AB 4700

Flow of data processing Fraction #1 Fraction #2 Fraction #3 Fraction #4 Fraction #5 db search (Mascot) Protein list #1 Protein list #2 Protein list #3 Protein list #4. . . non-redundant list of proteins non-significant proteins removed • Compile in Oracle db • generate quieries Fraction #1 Fraction #2 Fraction #3 Fraction #4 Fraction #5

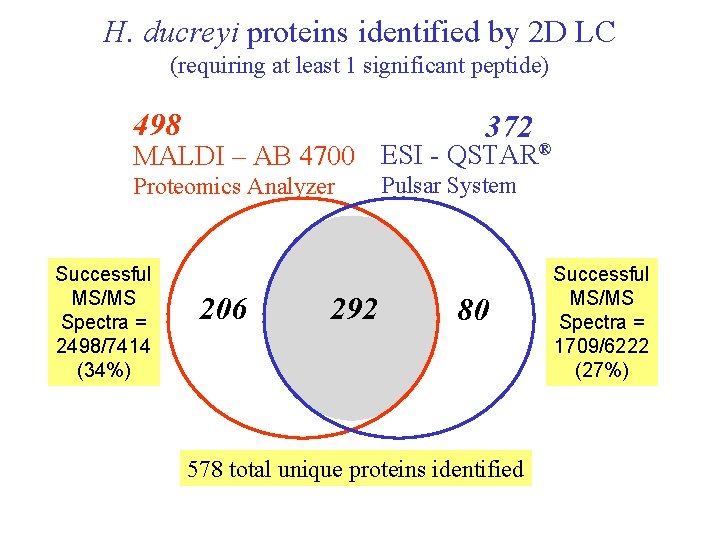
H. ducreyi proteins identified by 2 D LC (requiring at least 1 significant peptide) 498 372 MALDI – AB 4700 ESI - QSTAR® Proteomics Analyzer Successful MS/MS Spectra = 2498/7414 (34%) 206 292 Pulsar System 80 578 total unique proteins identified Successful MS/MS Spectra = 1709/6222 (27%)

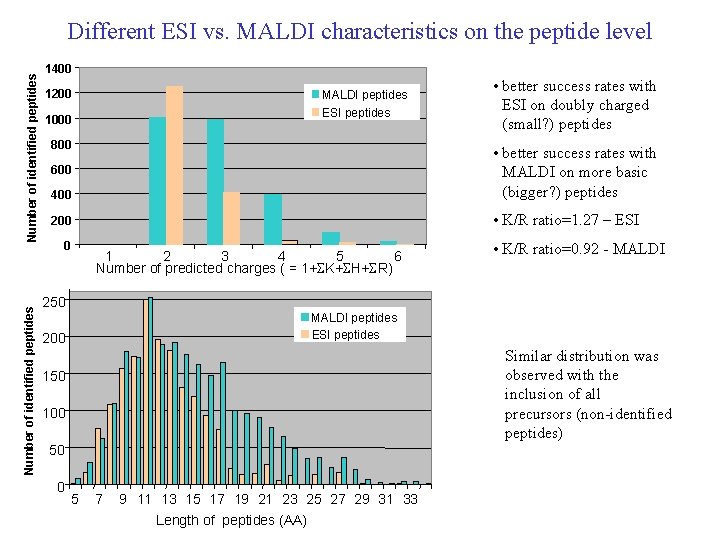
Number of identified peptides Different ESI vs. MALDI characteristics on the peptide level 1400 1200 MALDI peptides ESI peptides 1000 800 • better success rates with ESI on doubly charged (small? ) peptides 400 • better success rates with MALDI on more basic (bigger? ) peptides 200 • K/R ratio=1. 27 – ESI 600 0 1 2 3 4 5 6 Number of predicted charges ( = 1+SK+SH+SR) • K/R ratio=0. 92 - MALDI 250 MALDI peptides ESI peptides 200 Similar distribution was observed with the inclusion of all precursors (non-identified peptides) 150 100 50 0 5 7 9 11 13 15 17 19 21 23 25 27 29 31 33 Length of peptides (AA)

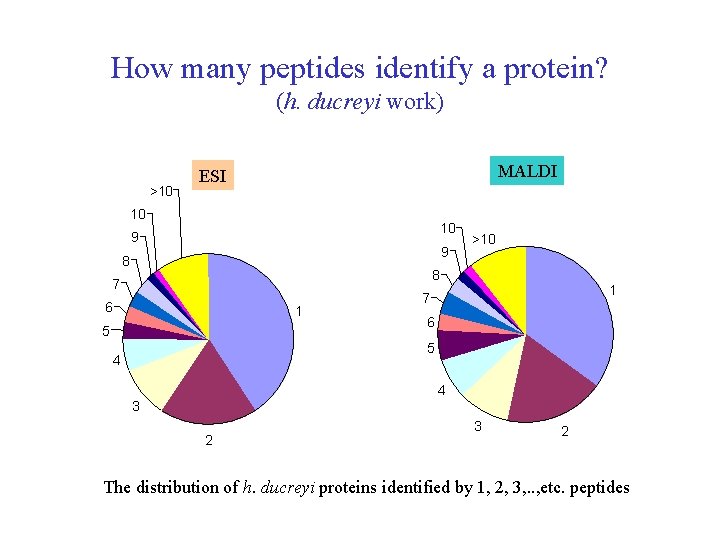
How many peptides identify a protein? (h. ducreyi work) >10 MALDI ESI 10 10 9 9 8 >10 8 7 6 1 5 1 7 6 5 4 4 3 2 The distribution of h. ducreyi proteins identified by 1, 2, 3, . . , etc. peptides

Conclusions from h. ducreyi work • From very complex mixtures MALDI had better efficiency of identification (higher “MS/MS duty cycle”) • Smaller peptides with K C-terminus identified more efficiently with ESI • Larger/more basic peptides are more efficiently identified by MALDI • A more complete sequence coverage of proteins is expected to “smooth out” differences

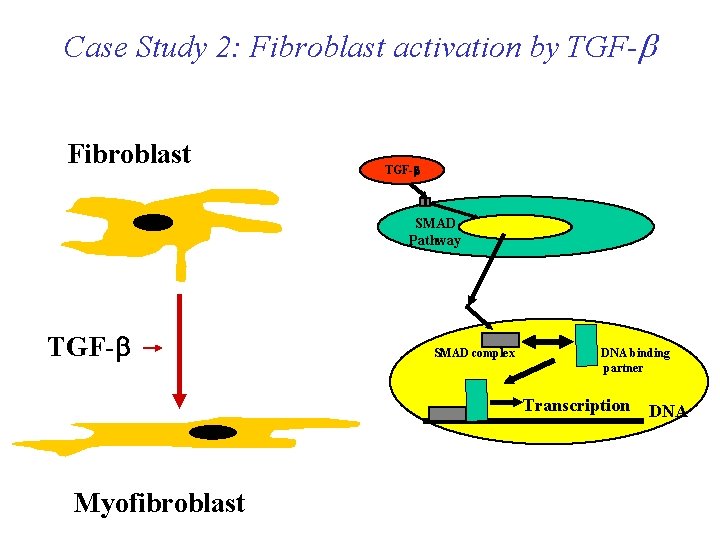
Case Study 2: Fibroblast activation by TGF-b Fibroblast TGF-b SMAD Pathway TGF-b SMAD complex DNA binding partner Transcription Myofibroblast DNA

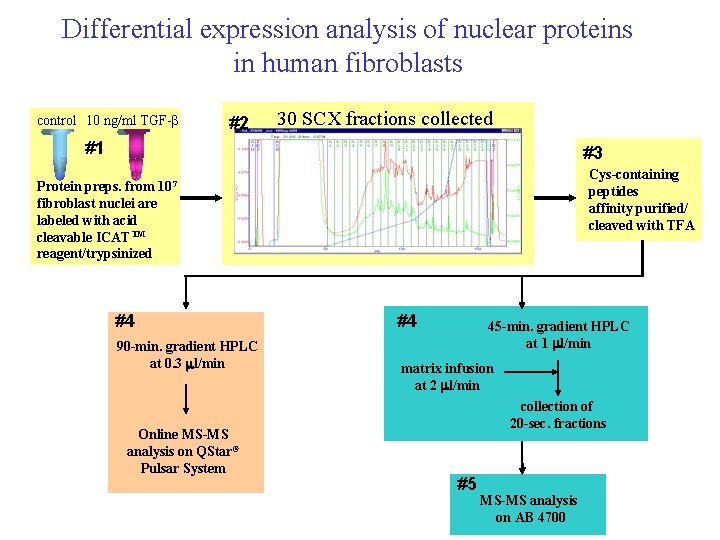
Differential expression analysis of nuclear proteins in human fibroblasts control 10 ng/ml TGF-b #2 30 SCX fractions collected #1 #3 Cys-containing peptides affinity purified/ cleaved with TFA Protein preps. from 107 fibroblast nuclei are labeled with acid cleavable ICATTM reagent/trypsinized #4 90 -min. gradient HPLC at 0. 3 ml/min Online MS-MS analysis on QStar® Pulsar System #4 45 -min. gradient HPLC at 1 ml/min matrix infusion at 2 ml/min collection of 20 -sec. fractions #5 MS-MS analysis on AB 4700

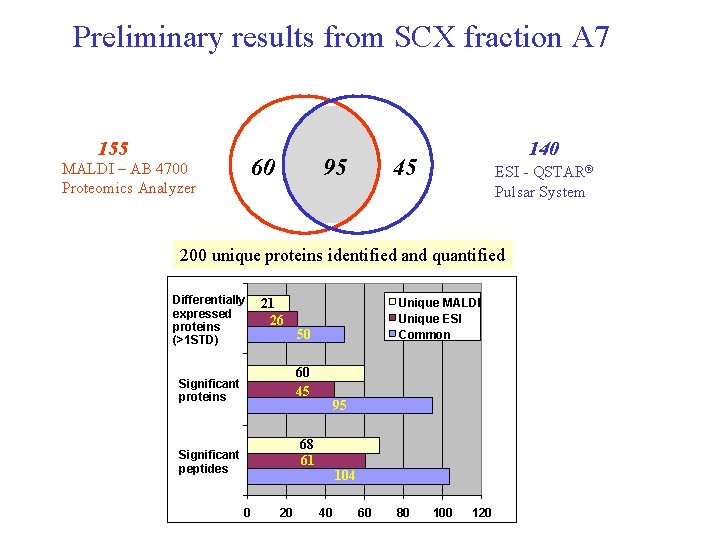
Preliminary results from SCX fraction A 7 155 60 MALDI – AB 4700 Proteomics Analyzer 95 140 45 ESI - QSTAR® Pulsar System 200 unique proteins identified and quantified Differentially expressed proteins (>1 STD) 21 26 Unique MALDI Unique ESI Common 50 60 45 Significant proteins 95 68 61 Significant peptides 104 0 20 40 60 80 100 120

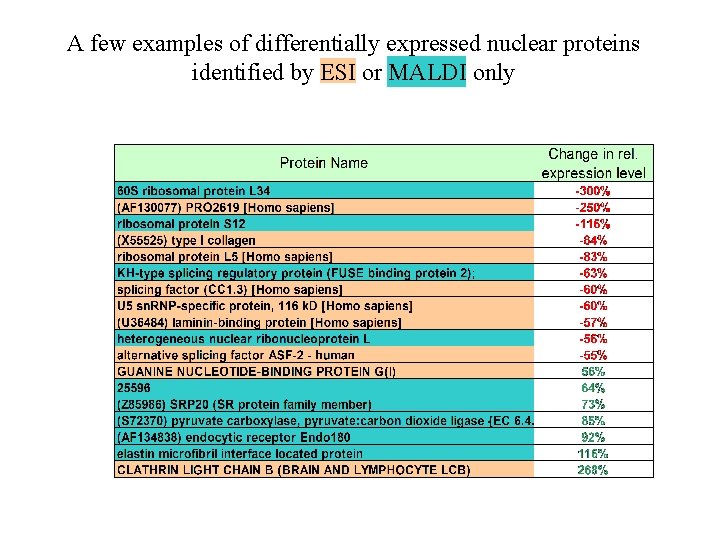
A few examples of differentially expressed nuclear proteins identified by ESI or MALDI only

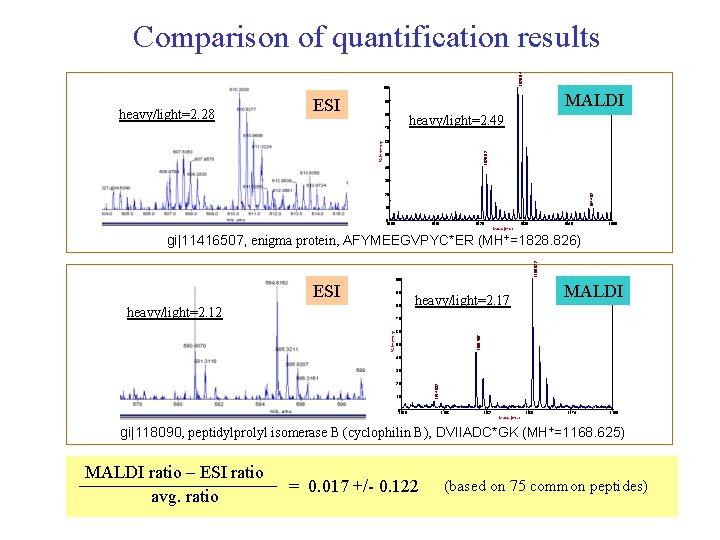
1828. 84 Comparison of quantification results 100 heavy/light=2. 28 ESI 1419. 6 MALDI 90 80 heavy/light=2. 49 60 1820. 82 % Intensity 70 50 40 30 1844. 82 20 10 0 1800 1810 1820 1830 1840 1850 Mass (m/z) 1168. 622 gi|11416507, enigma protein, AFYMEEGVPYC*ER (MH+=1828. 826) 100 ESI 90 80 heavy/light=2. 12 1502. 7 MALDI heavy/light=2. 17 60 1160. 597 % Intensity 70 50 40 20 10 0 1154. 632 30 1156 1162 1168 1174 1180 Mass (m/z) gi|118090, peptidylprolyl isomerase B (cyclophilin B), DVIIADC*GK (MH+=1168. 625) MALDI ratio – ESI ratio avg. ratio = 0. 017 +/- 0. 122 (based on 75 common peptides)

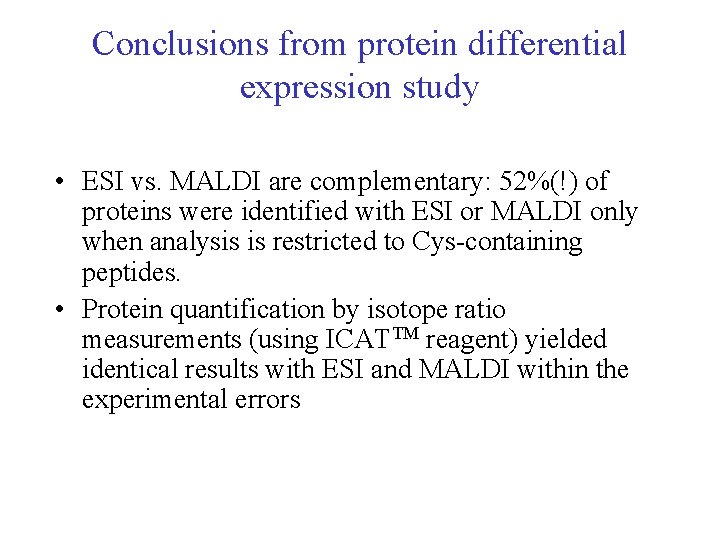
Conclusions from protein differential expression study • ESI vs. MALDI are complementary: 52%(!) of proteins were identified with ESI or MALDI only when analysis is restricted to Cys-containing peptides. • Protein quantification by isotope ratio measurements (using ICATTM reagent) yielded identical results with ESI and MALDI within the experimental errors