Enzymology Lecture 3 and 4 by Rumeza Hanif

Enzymology Lecture 3 and 4 by Rumeza Hanif
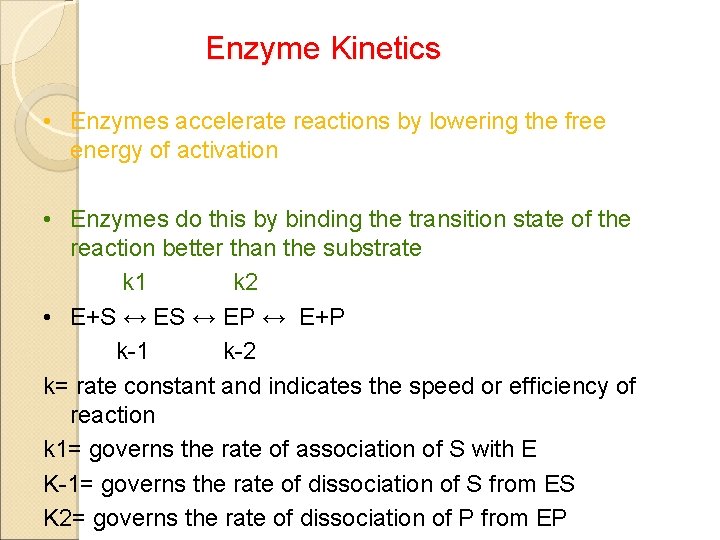
Enzyme Kinetics • Enzymes accelerate reactions by lowering the free energy of activation • Enzymes do this by binding the transition state of the reaction better than the substrate k 1 k 2 • E+S ↔ EP ↔ E+P k-1 k-2 k= rate constant and indicates the speed or efficiency of reaction k 1= governs the rate of association of S with E K-1= governs the rate of dissociation of S from ES K 2= governs the rate of dissociation of P from EP

The Michaelis-Menten Equation Louis Michaelis and Maude Menten's theory It assumes the formation of an enzymesubstrate complex It assumes that the ES complex is in rapid equilibrium with free enzyme Breakdown of ES to form products is assumed to be slower than (1) formation of ES and (2) breakdown of ES to re-form E and S

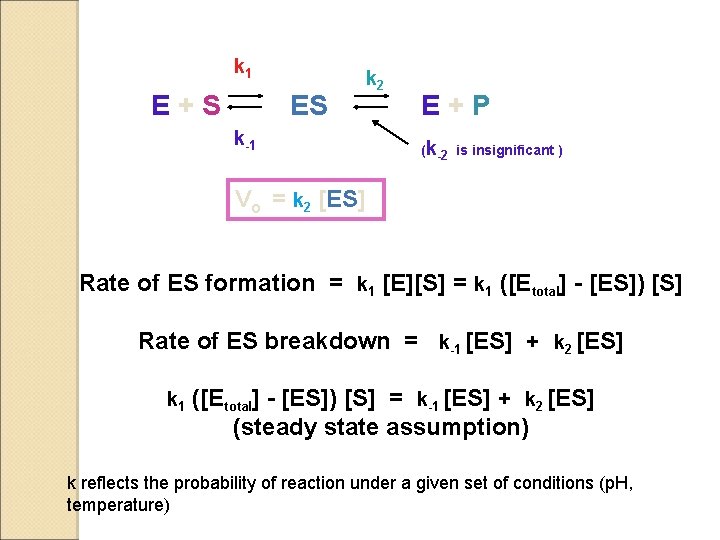
k 1 E+S ES k 2 k-1 E+P (k-2 is insignificant ) Vo = k 2 [ES] Rate of ES formation = k 1 [E][S] = k 1 ([Etotal] - [ES]) [S] Rate of ES breakdown = k-1 [ES] + k 2 [ES] k 1 ([Etotal] - [ES]) [S] = k-1 [ES] + k 2 [ES] (steady state assumption) k reflects the probability of reaction under a given set of conditions (p. H, temperature)
![k 1 [Etotal][S] - k 1[ES][S] = ( k-1 + k 2 )[ES] k k 1 [Etotal][S] - k 1[ES][S] = ( k-1 + k 2 )[ES] k](http://slidetodoc.com/presentation_image_h/12b4e3a5e77d891751a41a964456fbd9/image-6.jpg)
k 1 [Etotal][S] - k 1[ES][S] = ( k-1 + k 2 )[ES] k 1 [Etotal][S] = (k 1[S] + k-1 + k 2 )[ES] = [Etotal][S] ____________ = [S] + (k 2 + k-1 ) ______ k 1 Vo = k 2 [ES] Vo = Vmax when [Etotal] = [ES] (at saturation) Vo = [Etotal][S] ______ KM + [S] k 2 [Etotal][S] ______ KM + [S] Therefore Vmax = k 2 [Etotal] Vmax[S] ______ KM + [S]
![The dual nature of the Michaelis-Menten equation Vo = Vmax[S] _____ Km + [S] The dual nature of the Michaelis-Menten equation Vo = Vmax[S] _____ Km + [S]](http://slidetodoc.com/presentation_image_h/12b4e3a5e77d891751a41a964456fbd9/image-7.jpg)
The dual nature of the Michaelis-Menten equation Vo = Vmax[S] _____ Km + [S] Combination of zero-order and first-order kinetics When [S] is low, the equation for rate is first order in [S] When [S] is high, the equation for rate is zero-order in [S] The Michaelis-Menten equation describes a rectangular hyperbolic dependence of Vo on [S]
![Enzyme Kinetics: Michaelis-Menton Equation Vo = Vmax[S] ______ KM + [S] KM = [S] Enzyme Kinetics: Michaelis-Menton Equation Vo = Vmax[S] ______ KM + [S] KM = [S]](http://slidetodoc.com/presentation_image_h/12b4e3a5e77d891751a41a964456fbd9/image-8.jpg)
Enzyme Kinetics: Michaelis-Menton Equation Vo = Vmax[S] ______ KM + [S] KM = [S] when Vo = Vmax _____ 2 From Lehninger Principles of Biochemistry
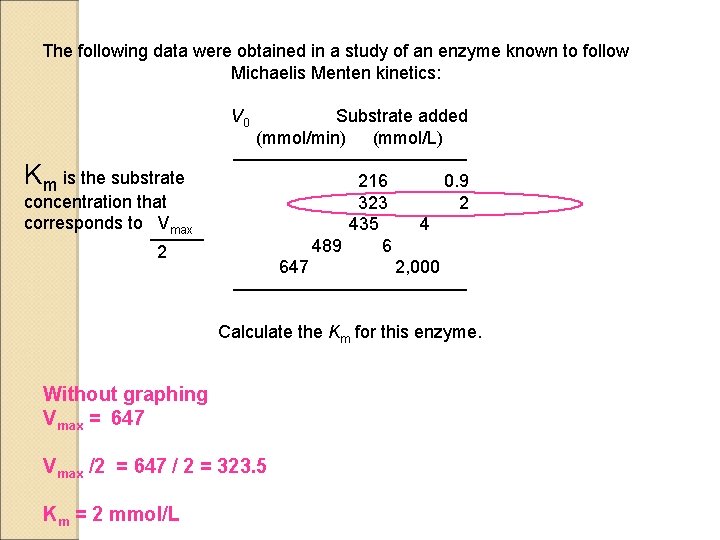
The following data were obtained in a study of an enzyme known to follow Michaelis Menten kinetics: V 0 Km is the substrate concentration that corresponds to Vmax 2 Substrate added (mmol/min) (mmol/L) ——————— 216 0. 9 323 2 435 4 489 6 647 2, 000 ——————— Calculate the Km for this enzyme. Without graphing Vmax = 647 Vmax /2 = 647 / 2 = 323. 5 Km = 2 mmol/L
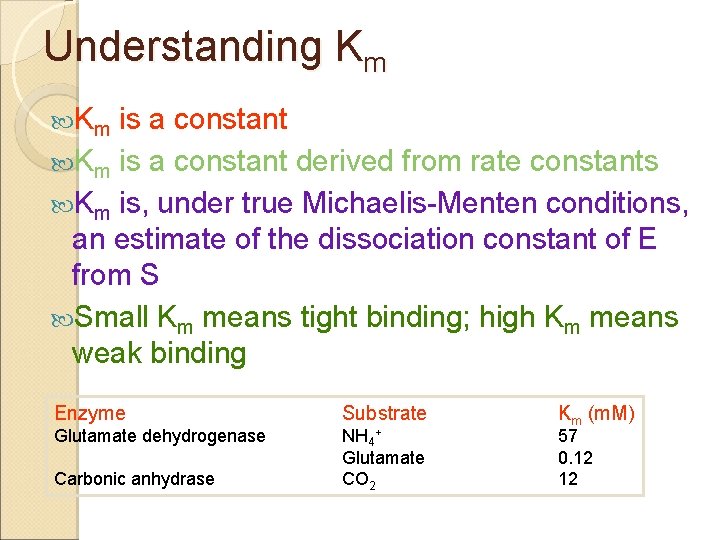
Understanding Km Km is a constant derived from rate constants Km is, under true Michaelis-Menten conditions, an estimate of the dissociation constant of E from S Small Km means tight binding; high Km means weak binding Enzyme Substrate Glutamate dehydrogenase NH 4+ Glutamate CO 2 Carbonic anhydrase Km (m. M) 57 0. 12 12

Understanding Vmax The theoretical maximal velocity Vmax is a constant Vmax is theoretical maximal rate of the reaction - but it is NEVER achieved in reality To reach Vmax would require that ALL enzyme molecules are tightly bound with substrate Vmax is asymptotically approached as substrate is increased

The turnover number (also known as the molecular activity of the enzyme) A measure of its maximal catalytic activity At high substrate concentration the overall velocity of the reaction is Vmax and the rate is determined by the enzyme concentration. The rate constant observed under these conditions is called the catalytic constant. kcat, the turnover number, is the number of substrate molecules converted to product per enzyme molecule per unit of time, when E is saturated with substrate. It measures how quickly a given enzyme can catalyze a specific reaction. If the M-M model fits, k 2 = kcat = Vmax/Et Values of kcat range from less than 1/sec to many millions per sec
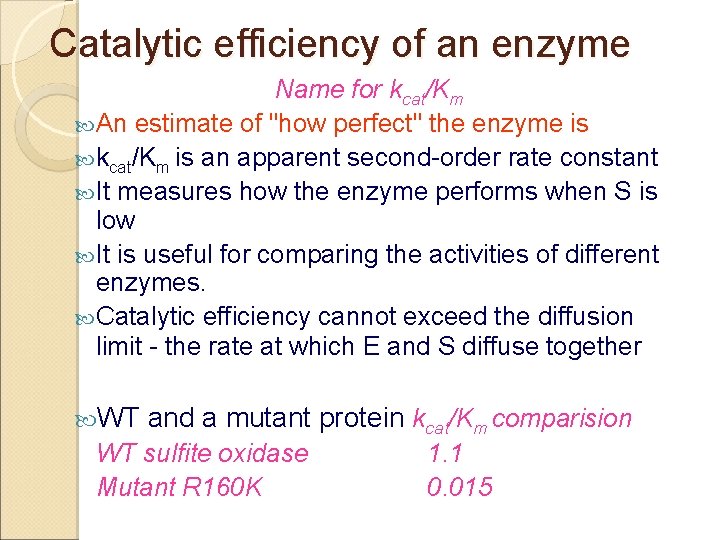
Catalytic efficiency of an enzyme Name for kcat/Km An estimate of "how perfect" the enzyme is kcat/Km is an apparent second-order rate constant It measures how the enzyme performs when S is low It is useful for comparing the activities of different enzymes. Catalytic efficiency cannot exceed the diffusion limit - the rate at which E and S diffuse together WT and a mutant protein kcat/Km comparision WT sulfite oxidase Mutant R 160 K 1. 1 0. 015
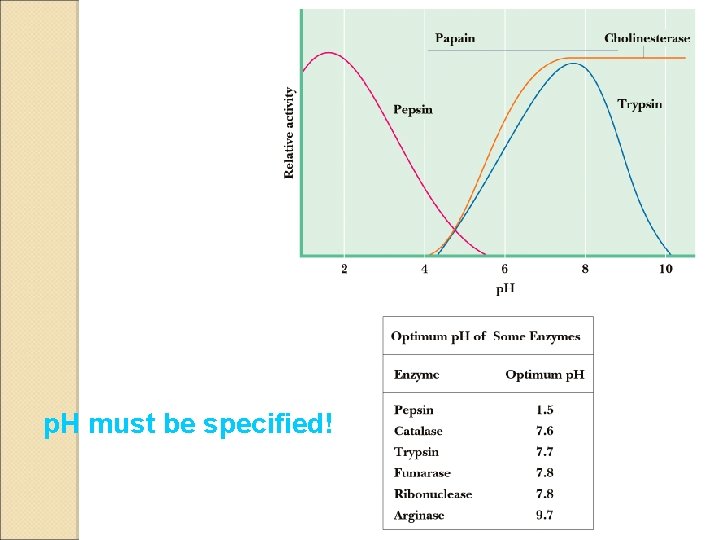
p. H must be specified!
Enzyme Inhibitors Reversible versus Irreversible Reversible inhibitors interact with an enzyme via noncovalent associations Irreversible inhibitors interact with an enzyme via covalent associations

Classes of Inhibition Two real, one hypothetical Competitive inhibition - inhibitor (I) binds only to E, not to ES Uncompetitive inhibition - inhibitor (I) binds only to ES, not to E. This is a hypothetical case that has never been documented for a real enzyme, but which makes a useful contrast to competitive inhibition Noncompetitive (mixed) inhibition - inhibitor (I) binds to E and to ES
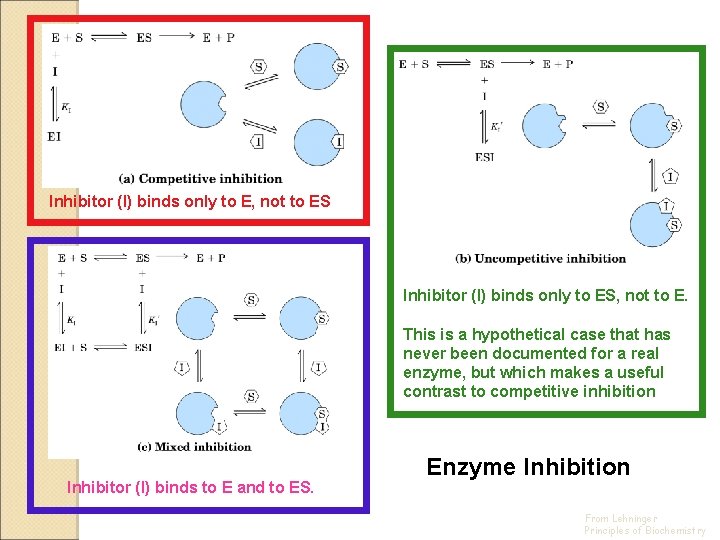
Inhibitor (I) binds only to E, not to ES Inhibitor (I) binds only to ES, not to E. This is a hypothetical case that has never been documented for a real enzyme, but which makes a useful contrast to competitive inhibition Inhibitor (I) binds to E and to ES. Enzyme Inhibition From Lehninger Principles of Biochemistry
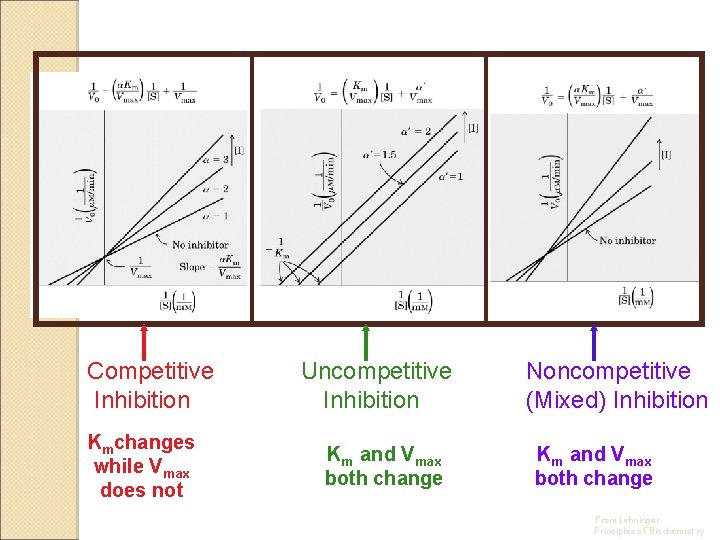
Competitive Inhibition Kmchanges while Vmax does not Uncompetitive Inhibition Km and Vmax both change Noncompetitive (Mixed) Inhibition Km and Vmax both change From Lehninger Principles of Biochemistry
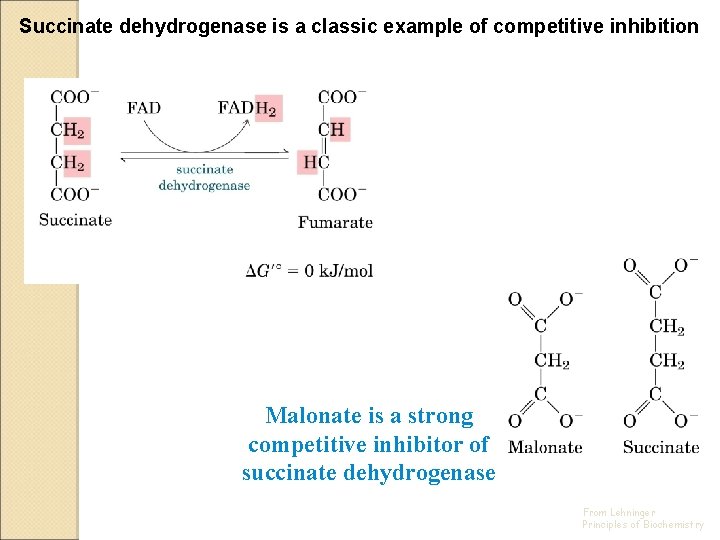
Succinate dehydrogenase is a classic example of competitive inhibition Malonate is a strong competitive inhibitor of succinate dehydrogenase From Lehninger Principles of Biochemistry
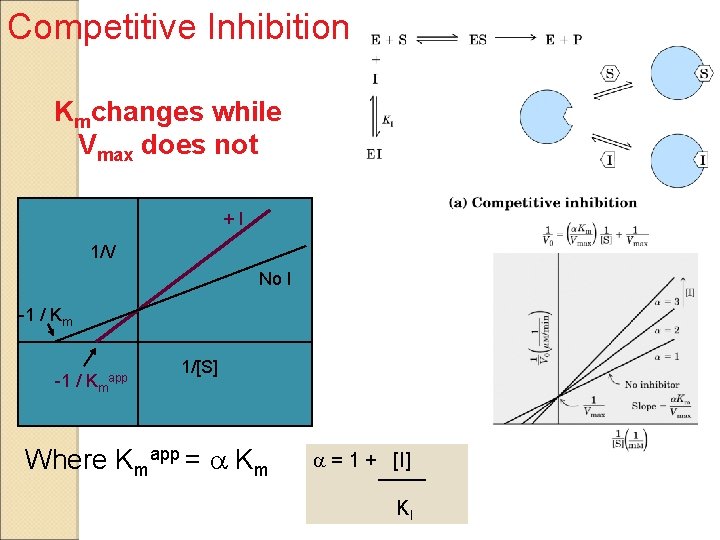
Competitive Inhibition Kmchanges while Vmax does not +I 1/V No I -1 / Kmapp 1/[S] Where Kmapp = a Km a = 1 + [I] KI
![Uncompetitive inhibition 1/V a’/Vmax +I No I -a’ / Km 1/Vmax 1/[S] -1 / Uncompetitive inhibition 1/V a’/Vmax +I No I -a’ / Km 1/Vmax 1/[S] -1 /](http://slidetodoc.com/presentation_image_h/12b4e3a5e77d891751a41a964456fbd9/image-22.jpg)
Uncompetitive inhibition 1/V a’/Vmax +I No I -a’ / Km 1/Vmax 1/[S] -1 / Km a‘ = 1 + [I] KI
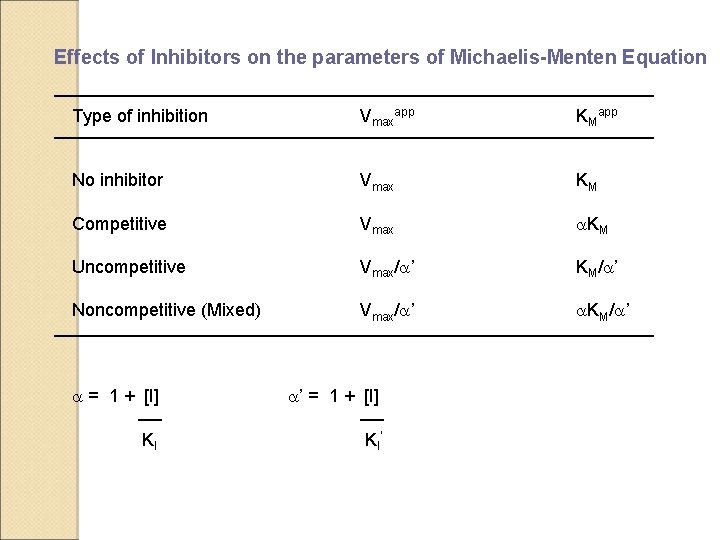
Effects of Inhibitors on the parameters of Michaelis-Menten Equation Type of inhibition Vmaxapp KMapp No inhibitor Vmax KM Competitive Vmax a. KM Uncompetitive Vmax/a’ KM/a’ Noncompetitive (Mixed) Vmax/a’ a. KM/a’ a = 1 + [I] a’ = 1 + [I] KI KI’

Reasoning? ? Reference: Chapter 5: properties of enzymes Principles of biochemistry, 4 th Ed, Pearson International Ed
Regulation of enzymatic activity Two ways that this may occur: 1) Control of enzyme availability Depends on rate of enzyme synthesis & degradation 2) Control of enzyme activity Enzyme-substrate binding affinity may vary with binding of small molecules called allosteric effectors (ex: BPG for Hb) Allosteric mechanisms can cause large changes in enzymatic activity
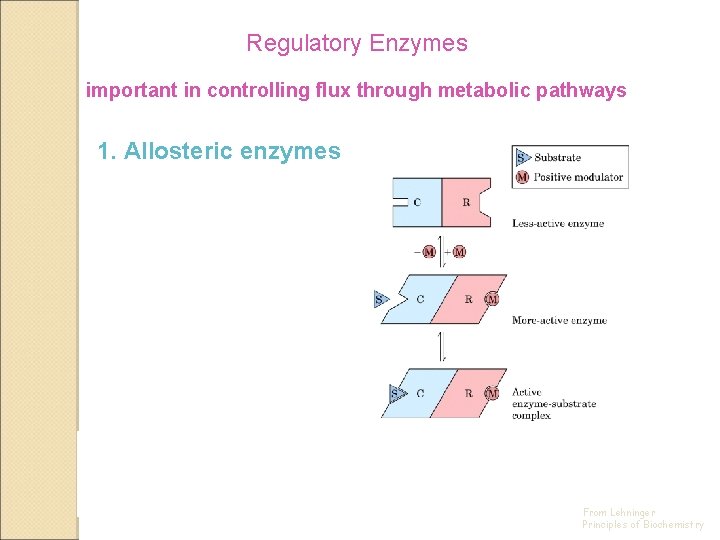
Regulatory Enzymes important in controlling flux through metabolic pathways 1. Allosteric enzymes 2. Regulation by covalent modification From Lehninger Principles of Biochemistry
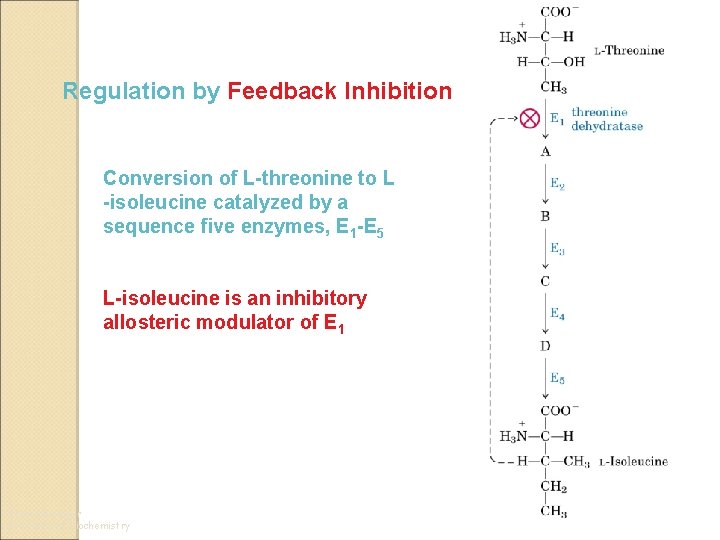
Regulation by Feedback Inhibition Conversion of L-threonine to L -isoleucine catalyzed by a sequence five enzymes, E 1 -E 5 L-isoleucine is an inhibitory allosteric modulator of E 1 From Lehninger Principles of Biochemistry
Multienzyme Complexes and Multifunctional Enzymes In some cases, different enzymes that catalyza sequential reactions in the same pathway are bound together in a multienzyme complex. In other cases, different activities can be found on a single multifunctional polypeptide chain.
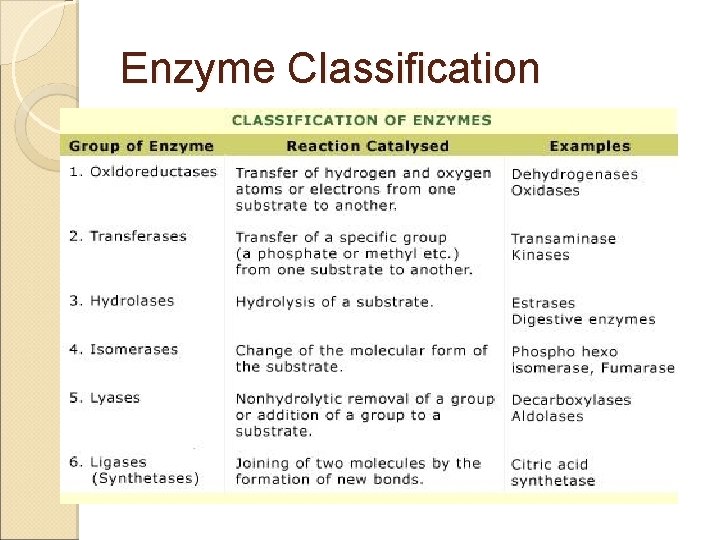
Enzyme Classification



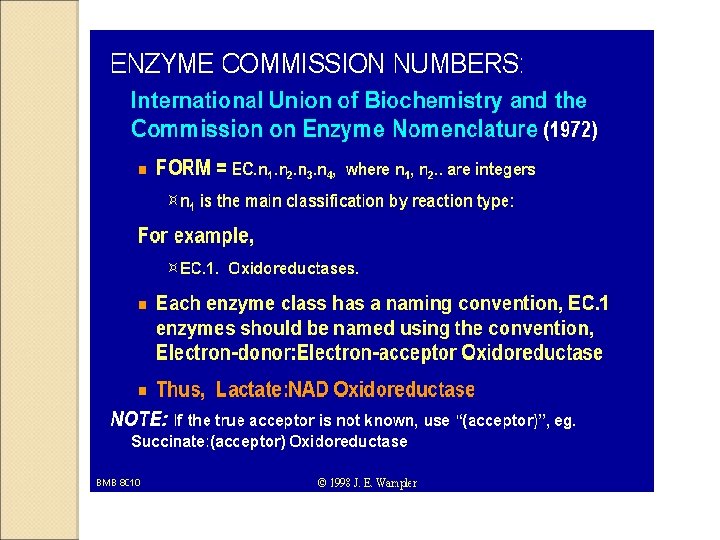

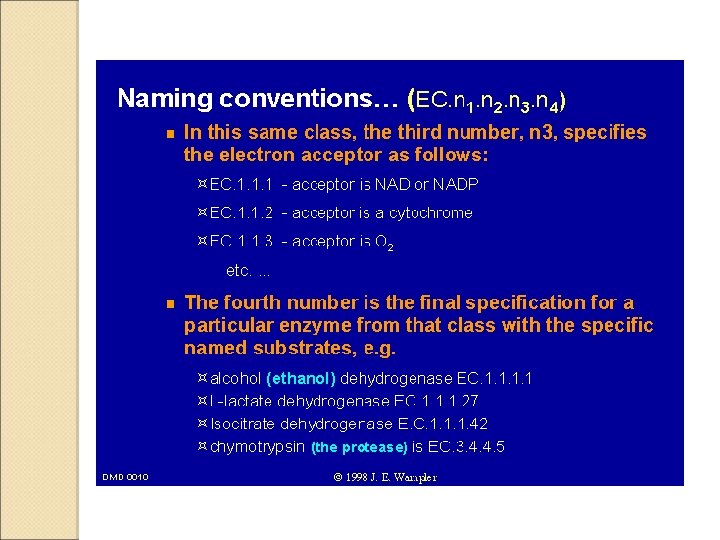
- Slides: 36