ENZYMES FOR MANIPULATING DNA Buffers and solution conditions

ENZYMES FOR MANIPULATING DNA *** Buffers and solution conditions*** I. DNA polymerases III. Kinase and alkaline phosphatase IV. Nucleases V. Topoisomerase

Buffers are crucial for activity of enzymes! Ideal biochemical buffers: • p. Ka between 6 and 8 • Chemically inert • Polar (soluble and not membrane permeable) • Non-toxic • Inexpensive • Salt and temperature indifferent
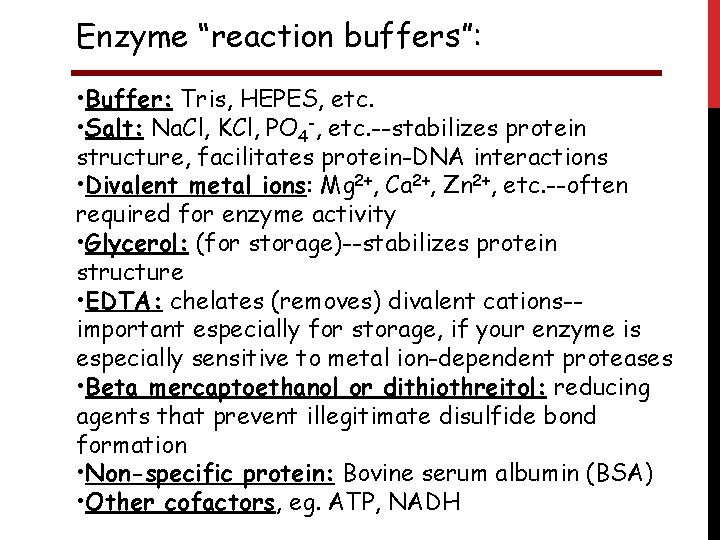
Enzyme “reaction buffers”: • Buffer: Tris, HEPES, etc. • Salt: Na. Cl, KCl, PO 4 -, etc. --stabilizes protein structure, facilitates protein-DNA interactions • Divalent metal ions: Mg 2+, Ca 2+, Zn 2+, etc. --often required for enzyme activity • Glycerol: (for storage)--stabilizes protein structure • EDTA: chelates (removes) divalent cations-important especially for storage, if your enzyme is especially sensitive to metal ion-dependent proteases • Beta mercaptoethanol or dithiothreitol: reducing agents that prevent illegitimate disulfide bond formation • Non-specific protein: Bovine serum albumin (BSA) • Other cofactors, eg. ATP, NADH
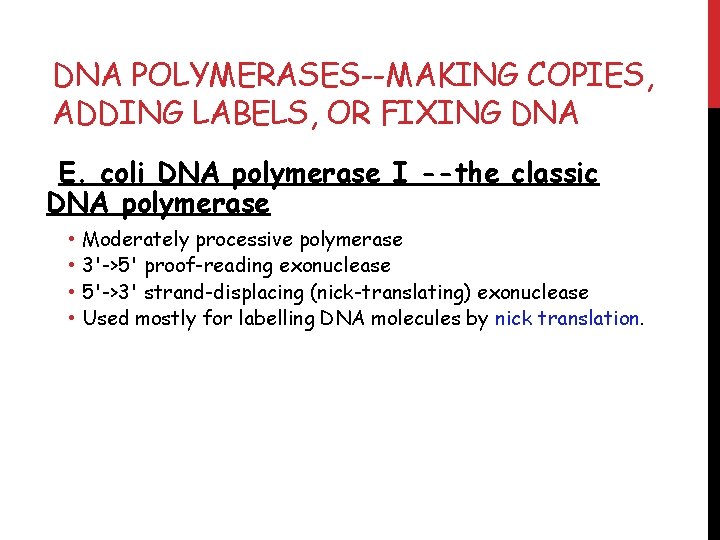
DNA POLYMERASES--MAKING COPIES, ADDING LABELS, OR FIXING DNA E. coli DNA polymerase I --the classic DNA polymerase • • Moderately processive polymerase 3'->5' proof-reading exonuclease 5'->3' strand-displacing (nick-translating) exonuclease Used mostly for labelling DNA molecules by nick translation.
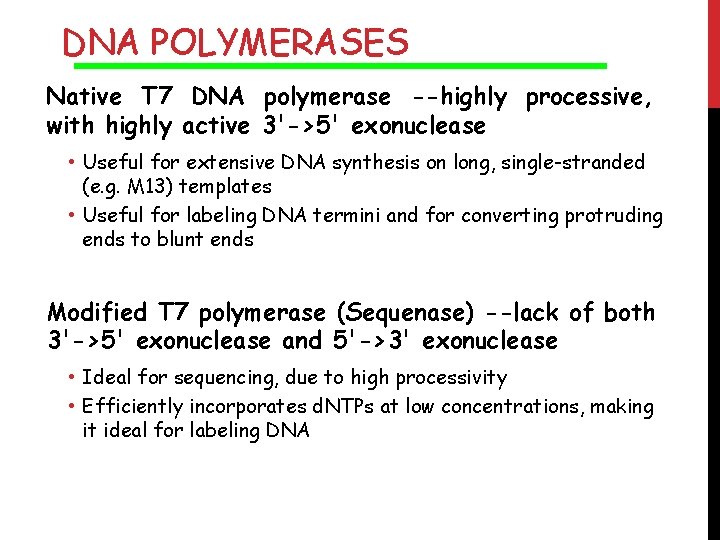
DNA POLYMERASES Native T 7 DNA polymerase --highly processive, with highly active 3'->5' exonuclease • Useful for extensive DNA synthesis on long, single-stranded (e. g. M 13) templates • Useful for labeling DNA termini and for converting protruding ends to blunt ends Modified T 7 polymerase (Sequenase) --lack of both 3'->5' exonuclease and 5'->3' exonuclease • Ideal for sequencing, due to high processivity • Efficiently incorporates d. NTPs at low concentrations, making it ideal for labeling DNA

DNA POLYMERASES Reverse transcriptase • RNA-dependent DNA polymerase • Essential for making c. DNA copies of RNA transcripts • Cloning intron-less genes • Quantitation of RNA
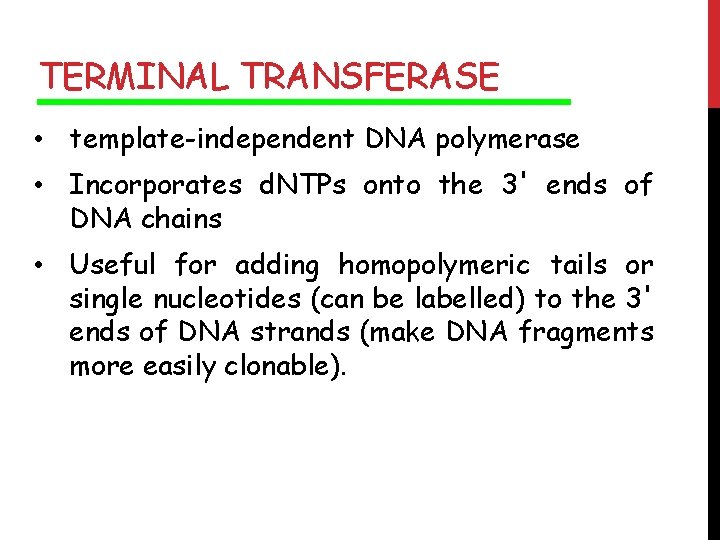
TERMINAL TRANSFERASE • template-independent DNA polymerase • Incorporates d. NTPs onto the 3' ends of DNA chains • Useful for adding homopolymeric tails or single nucleotides (can be labelled) to the 3' ends of DNA strands (make DNA fragments more easily clonable).
T 4 POLYNUCLEOTIDE KINASE • Transfers gamma phosphate of ATP to the 5’ end of polynucleotides • Useful for preparing DNA fragments for ligation (if they lack 5’ phosphates) • Useful for radiolabelling DNA fragments using gamma 32 P ATP as a phosphate donor
ALKALINE PHOSPHATASE • Catalyzes removal of 5’ (and 3’) phosphates from polynucleotides • Useful for treating restricted vector DNA sequences prior to ligation reactions, prevents religation of vector in the absence of insert DNA • Lack of vector 5’ phosphates may inhibit transformation efficiency? Use only when absolutely necessary…
NUCLEASES Exonucleases • Remove nucleotides one at a time from a DNA molecule Endonucleases • Break phosphodiester bonds within a DNA molecule • Include restriction enzymes

EXONUCLEASES Bal 31 • Double-stranded exonuclease, operates in a timedependent manner • Degrades both 5’ and 3’ ends of DNA • Useful for generating deletion sets, get bigger deletions with longer incubations.

EXONUCLEASES Exonuclease III--double-stranded DNA • 3’-5’ exonuclease activity • 3’ overhangs resistant to activity, can use this property to generate “nested” deletions from one end of a piece of DNA (use S 1 nuclease to degrade other strand of DNA)

EXONUCLEASES Exonuclease I • 3’-5’ exonuclease • Works only on single-stranded DNA • Useful for removing unextended primers from PCR reactions or other primer extension reactions

ENDONUCLEASES Dnase I • Cleaves double-stranded DNA randomly (also cleaves single -stranded DNA) • Mn++: both strands of DNA cut • Mg++: single strands nicked • Very useful for defining binding sites for DNA binding proteins
TYPES OF ENDONUCLEASES Type I: multisubunit proteins that function as a single protein complex, usually contain two R subunits, two M subunits and one S subunit Type II: recognize specific DNA sequences and cleave at constant positions at or close to that sequence to produce 5’-phosphates and 3’-hydroxyls. Most useful in cloning!! Type III: composed of two genes (mod and res) encoding protein subunits that function either in DNA recognition and modification (Mod) or restriction (Res) Type IV: one or two genes encoding proteins that cleave only modified DNA, including methylated, hydroxymethylated and glucosyl-hydroxymethylated bases

Topoisomerase • A restriction enzyme and ligase--all in one • altering the “linking number” in coiled, constrained (supercoiled) DNA--relaxing DNA twisting during replication
Topoisomerase • Topoisomerase catalyzed ligation is EXTREMELY efficient (>85% of resulting plasmids are recombinant)-excellent for library constructions • Can be used to clone blunt ended DNA (PCR products, restriction digests), T-overhang PCR products (from Taq polymerase), and directional clones
- Slides: 17