Enzyme Regulation Regulation of Enzyme Activity Enzyme quantity



![Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of](https://slidetodoc.com/presentation_image/c25bd9fa20274ebb20749678cff1f894/image-7.jpg)

- Slides: 14

Enzyme Regulation

Regulation of Enzyme Activity Enzyme quantity – regulation of gene expression (Response time = minutes to hours) a) Transcription b) Translation c) Enzyme turnover Enzyme activity (rapid response time = fraction of seconds) a) Allosteric regulation b) Covalent modification c) Association-disassociation’ d) Proteolytic cleavage of proenzyme

Allosteric Regulation • End products are often inhibitors • Allosteric modulators bind to site other than the active site • Allosteric enzymes usually have 4 o structure • Vo vs [S] plots give sigmoidal curve for at least one substrate • Can remove allosteric site without effecting enzymatic action

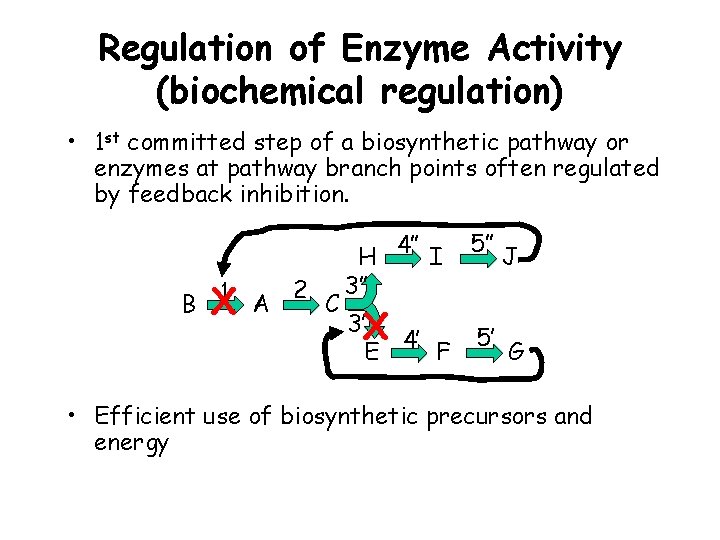
Regulation of Enzyme Activity (biochemical regulation) • 1 st committed step of a biosynthetic pathway or enzymes at pathway branch points often regulated by feedback inhibition. 1 A 2 C B X H 4” I 3” 3’X E 4’ F 5” 5’ J G • Efficient use of biosynthetic precursors and energy

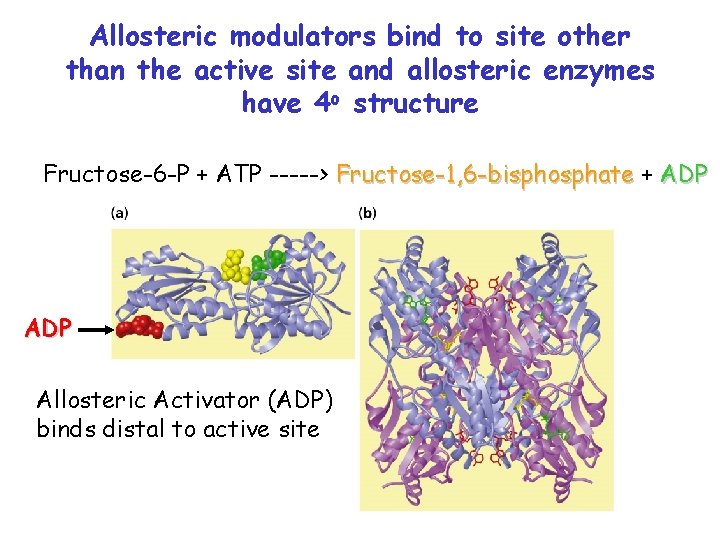
Phosphofructokinase( PFK) Fructose-6 -P + ATP -----> Fructose-1, 6 -bisphosphate + ADP • PFK catalyzes 1 st committed step in glycolysis (10 steps total) (Glucose + 2 ADP + 2 NAD+ + 2 Pi 2 pyruvate + 2 ATP + 2 NADH) • Phosphoenolpyruvate is an allosteric inhibitor of PFK • ADP is an allosteric activator of PFK

Allosteric modulators bind to site other than the active site and allosteric enzymes have 4 o structure Fructose-6 -P + ATP -----> Fructose-1, 6 -bisphosphate + ADP Allosteric Activator (ADP) binds distal to active site
![Vo vs S plots give sigmoidal curve for at least one substrate Binding of Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of](https://slidetodoc.com/presentation_image/c25bd9fa20274ebb20749678cff1f894/image-7.jpg)
Vo vs [S] plots give sigmoidal curve for at least one substrate Binding of allosteric inhibitor or activator does not effect Vmax, but does alter Km Allosteric enzyme do not follow M-M kinetics

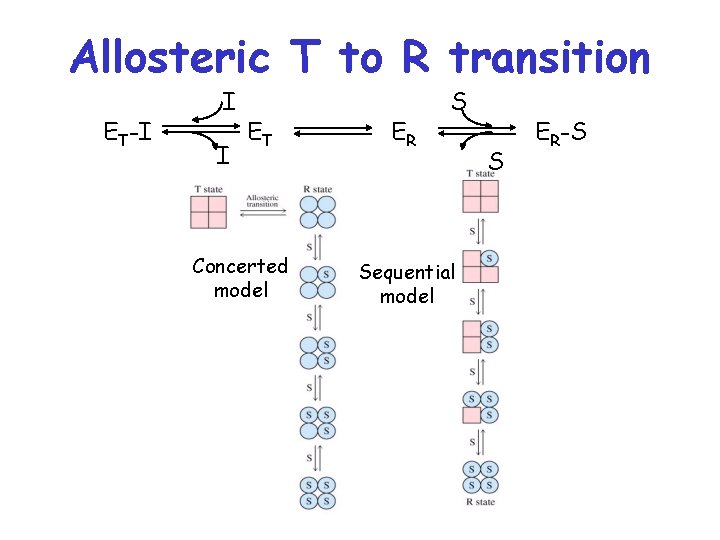
Allosteric T to R transition ET-I I I ET Concerted model ER S Sequential model S ER-S

Covalent modification • Regulation by covalent modification is slower than allosteric regulation • Reversible • Require one enzyme for activation and one enzyme for inactivation • Covalent modification freezes enzyme T or R conformation

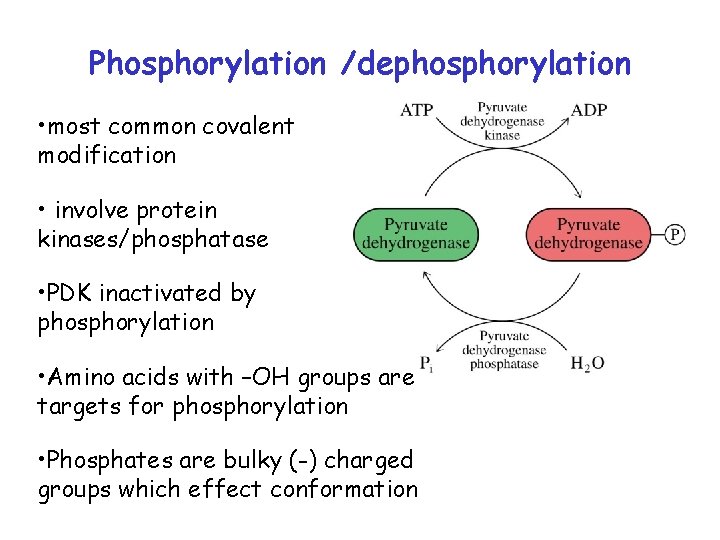
Phosphorylation /dephosphorylation • most common covalent modification • involve protein kinases/phosphatase • PDK inactivated by phosphorylation • Amino acids with –OH groups are targets for phosphorylation • Phosphates are bulky (-) charged groups which effect conformation

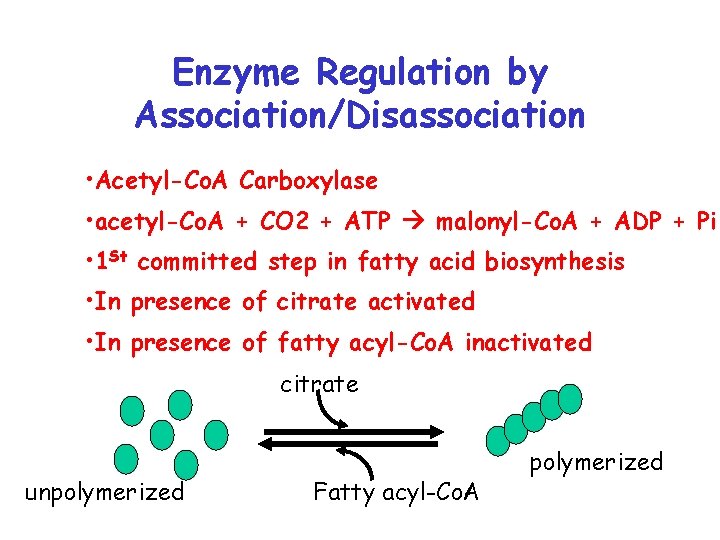
Enzyme Regulation by Association/Disassociation • Acetyl-Co. A Carboxylase • acetyl-Co. A + CO 2 + ATP malonyl-Co. A + ADP + Pi • 1 St committed step in fatty acid biosynthesis • In presence of citrate activated • In presence of fatty acyl-Co. A inactivated citrate unpolymerized Fatty acyl-Co. A polymerized

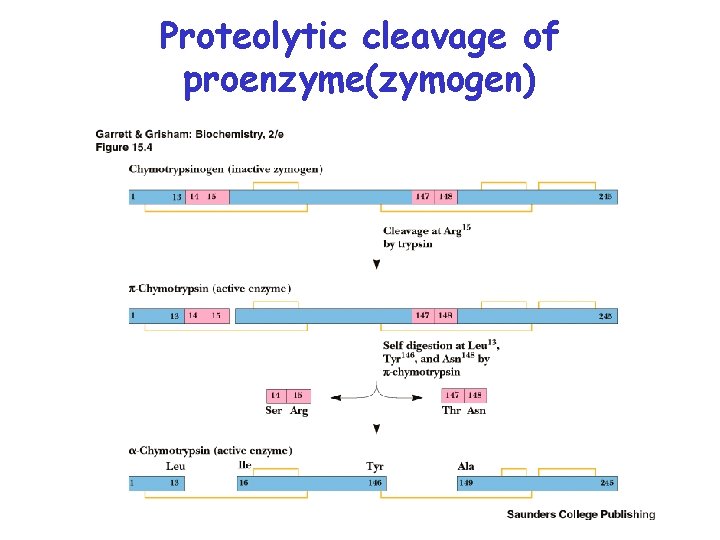
Proteolytic cleavage of proenzyme(zymogen)

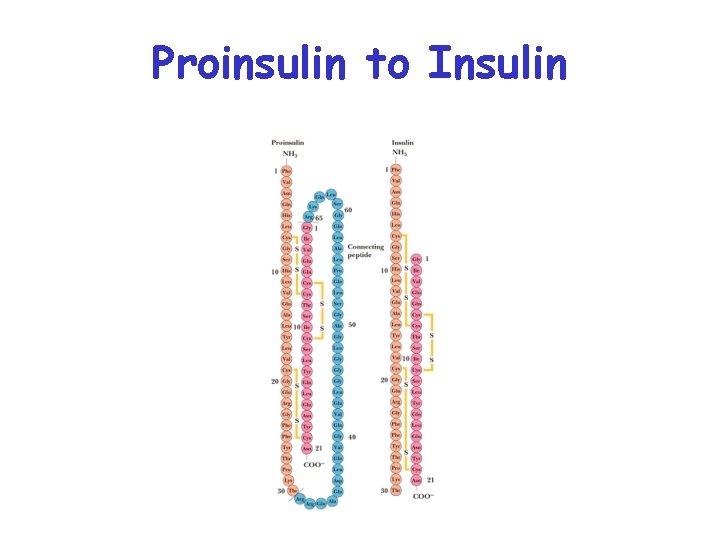
Proinsulin to Insulin

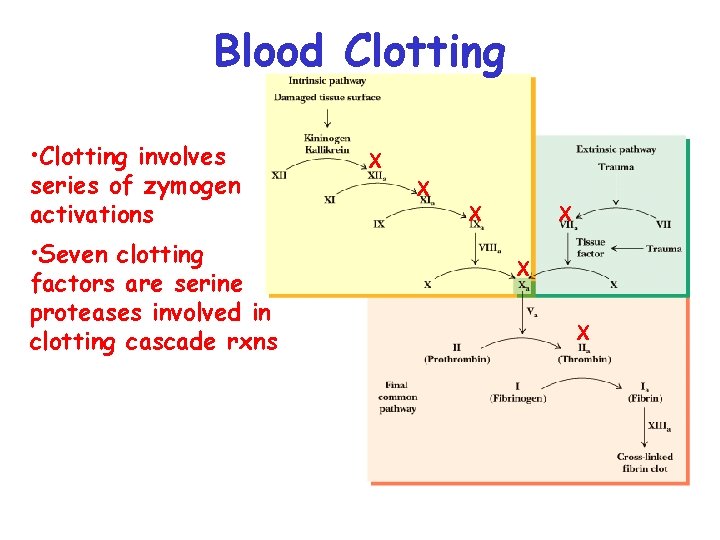
Blood Clotting • Clotting involves series of zymogen activations • Seven clotting factors are serine proteases involved in clotting cascade rxns X X X