Enumerate the differences between Lactose tryptophan operons Lac
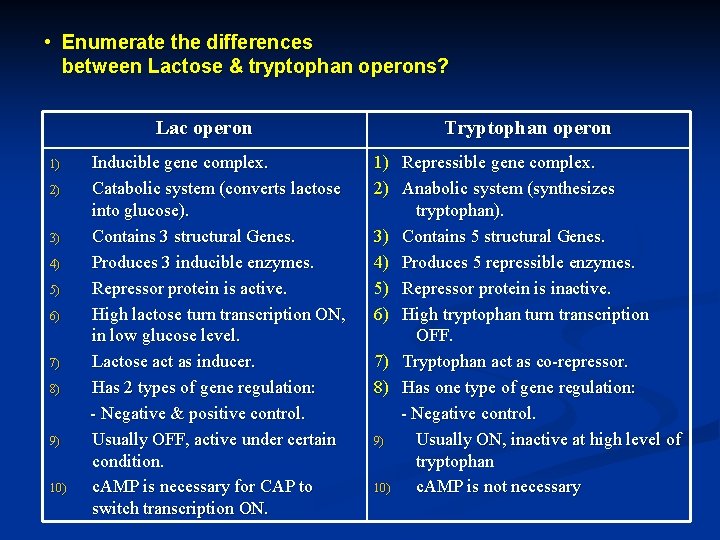
• Enumerate the differences between Lactose & tryptophan operons? Lac operon 1) 2) 3) 4) 5) 6) 7) 8) 9) 10) Inducible gene complex. Catabolic system (converts lactose into glucose). Contains 3 structural Genes. Produces 3 inducible enzymes. Repressor protein is active. High lactose turn transcription ON, in low glucose level. Lactose act as inducer. Has 2 types of gene regulation: - Negative & positive control. Usually OFF, active under certain condition. c. AMP is necessary for CAP to switch transcription ON. Tryptophan operon 1) Repressible gene complex. 2) Anabolic system (synthesizes tryptophan). 3) Contains 5 structural Genes. 4) Produces 5 repressible enzymes. 5) Repressor protein is inactive. 6) High tryptophan turn transcription OFF. 7) Tryptophan act as co-repressor. 8) Has one type of gene regulation: - Negative control. 9) Usually ON, inactive at high level of tryptophan 10) c. AMP is not necessary
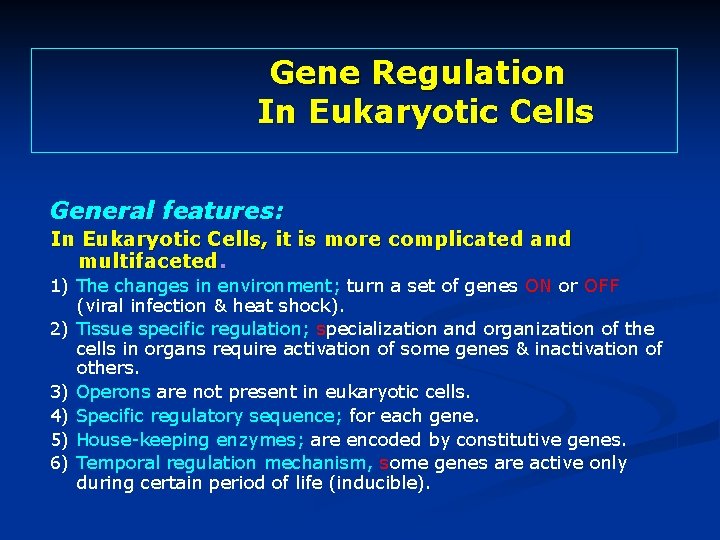
Gene Regulation In Eukaryotic Cells General features: In Eukaryotic Cells, it is more complicated and multifaceted. 1) The changes in environment; turn a set of genes ON or OFF (viral infection & heat shock). 2) Tissue specific regulation; specialization and organization of the cells in organs require activation of some genes & inactivation of others. 3) Operons are not present in eukaryotic cells. 4) Specific regulatory sequence; for each gene. 5) House-keeping enzymes; are encoded by constitutive genes. 6) Temporal regulation mechanism, some genes are active only during certain period of life (inducible).

Levels of Gene Expression Control In Eukaryotic Cells 1) 2) 3) 4) 5) DNA or Chromosomal organization. Gene transcription level. Post-transcription level. Translation control. Post-translation control.
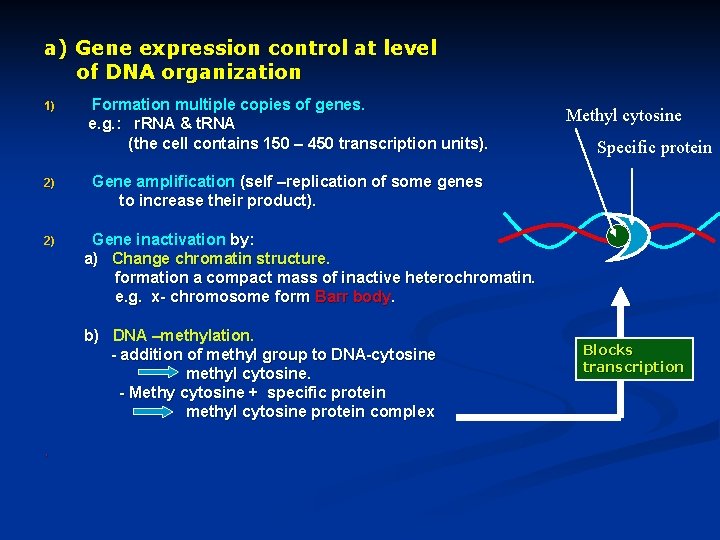
a) Gene expression control at level of DNA organization 1) 2) 2) Formation multiple copies of genes. e. g. : r. RNA & t. RNA (the cell contains 150 – 450 transcription units). Specific protein Gene amplification (self –replication of some genes to increase their product). Gene inactivation by: a) Change chromatin structure. formation a compact mass of inactive heterochromatin. e. g. x- chromosome form Barr body. b) DNA –methylation. - addition of methyl group to DNA-cytosine methyl cytosine. - Methy cytosine + specific protein methyl cytosine protein complex . Methyl cytosine Blocks transcription
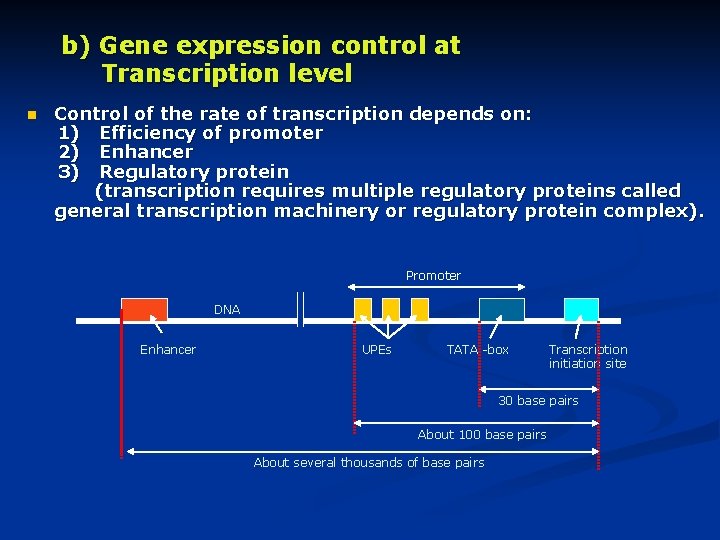
b) Gene expression control at Transcription level n Control of the rate of transcription depends on: 1) Efficiency of promoter 2) Enhancer 3) Regulatory protein (transcription requires multiple regulatory proteins called general transcription machinery or regulatory protein complex). Promoter DNA Enhancer UPEs TATA -box Transcription initiation site 30 base pairs About 100 base pairs About several thousands of base pairs
1) Promoter n Composition: 1) TATA –box : 2) UPEs (upstream promoter elements): n n Activity of promoter depends on the number & type of UPEs. Weak promoter contains few UPEs. Strong promoter contains several UPEs are required for accurate & efficient initiation of m. RNA – synthesis. • Binding site of RNA –polymerase. • Formed of T & A. • About 30 base pairs in upstream from transcription initiation site. • Each one about 8 – 12 base pairs • About 100 base pairs in upstream from initiation site.
2) Enhancer • • DNA –sequence Increase rate of Gene transcription, when it interacts with transcription machinery complex in presence of the activator protein 3) Regulatory protein • • • May be activator or repressor protein. Transcription requires multiple regulatory proteins, R. Protein complex. Regulatory protein complex binds to TATA –box to facilitate the binding of RNA polymerase, that results in formation of transcription machinery complex. 4) Activator protein • • Have 2 functional domains; one binds to Enhancer & the other connects the transcription machinery complex. Increase rate of transcription & m. RNA synthesis.
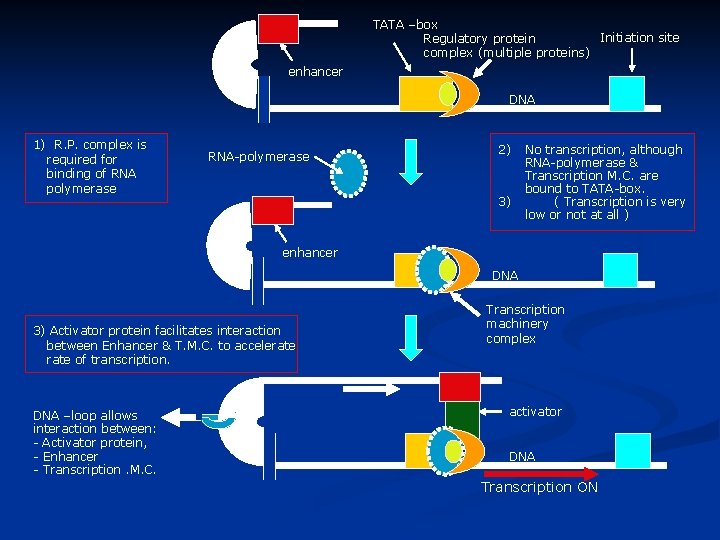
TATA –box Initiation site Regulatory protein complex (multiple proteins) enhancer DNA 1) R. P. complex is required for binding of RNA polymerase RNA-polymerase 2) 3) No transcription, although RNA-polymerase & Transcription M. C. are bound to TATA-box. ( Transcription is very low or not at all ) enhancer DNA 3) Activator protein facilitates interaction between Enhancer & T. M. C. to accelerate of transcription. DNA –loop allows interaction between: - Activator protein, - Enhancer - Transcription. M. C. Transcription machinery complex activator DNA Transcription ON
Gene Expression at Posttranslation level 1) Proteolytic process: Pepsinogen (inactive) Proteolytic enzymes Pepsin (active) 2) Selective degradation: To maintain a suitable concentration of protein within the cell and removal the unuseful parts. 3) Chemical modification: removal or addition functional group to activate or inactivate some enzymes, such as phosphate group.
- Slides: 9