ENGN 2500 Medical Image Analysis Project Final Presentation





- Slides: 30

ENGN 2500 Medical Image Analysis Project Final Presentation Extraction and Visualization of Humerus, Radius and Ulna of Dogs “Subvoxel Polygonization of Discrete Implicit Surfaces Using ENO Interpolation” Firat Kalaycilar

Problem Elbow dysplasia is a condition involving multiple developmental abnormalities of the elbow-joint in the dog, specifically the growth of cartilage or the structures surrounding it (from Wikipedia). The disease can be diagnosed using 3 D reconstruction of bones from CT data. Goal: Extract and visualize bones of interest in 3 D.


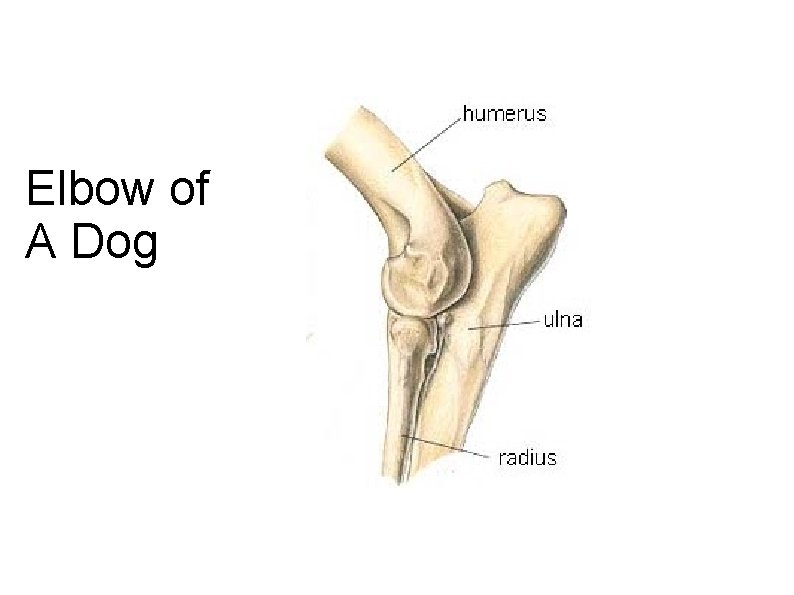
Elbow of A Dog

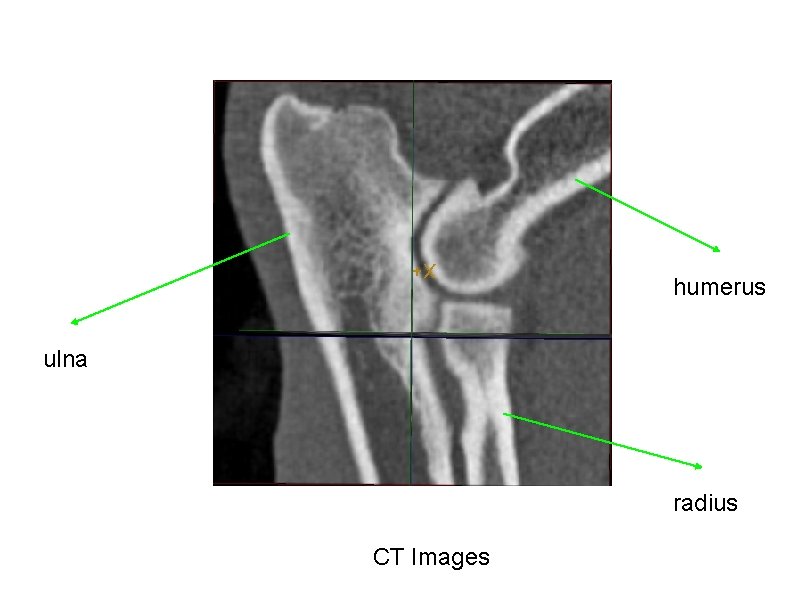
humerus ulna radius CT Images

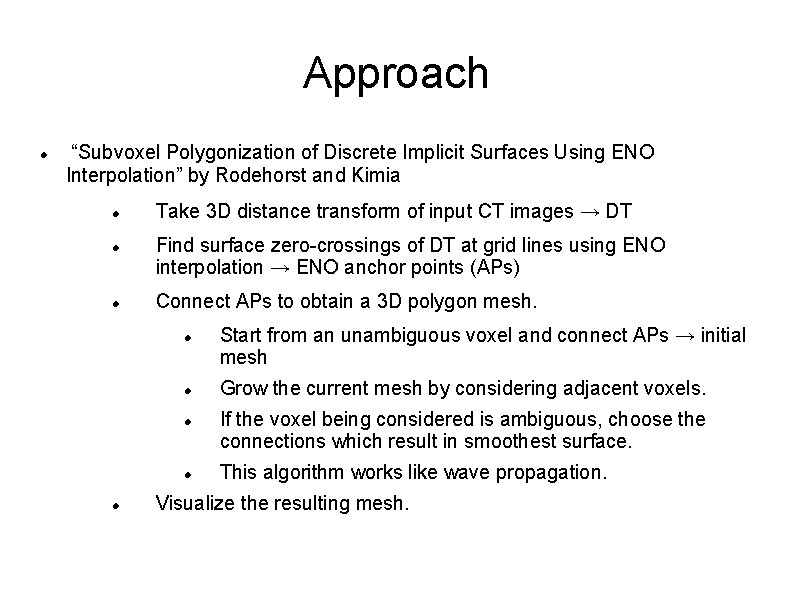
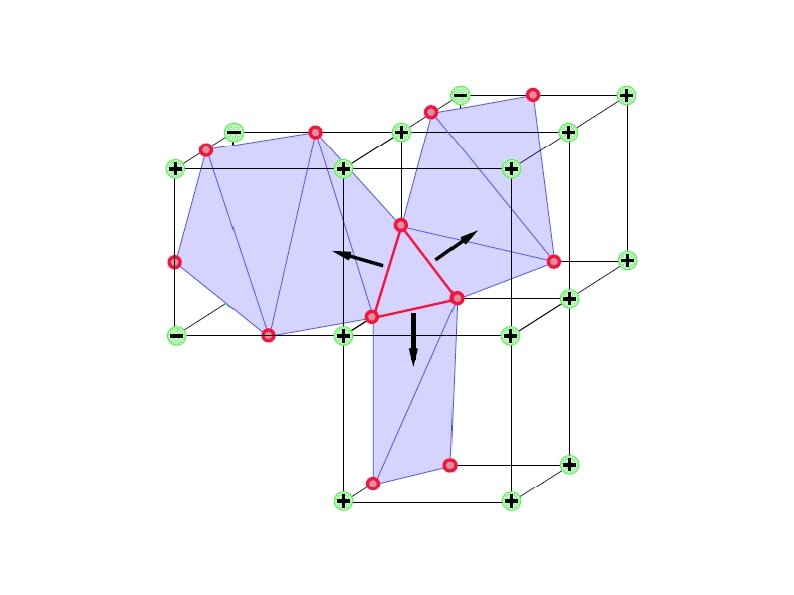
Approach “Subvoxel Polygonization of Discrete Implicit Surfaces Using ENO Interpolation” by Rodehorst and Kimia Take 3 D distance transform of input CT images → DT Find surface zero-crossings of DT at grid lines using ENO interpolation → ENO anchor points (APs) Connect APs to obtain a 3 D polygon mesh. Start from an unambiguous voxel and connect APs → initial mesh Grow the current mesh by considering adjacent voxels. If the voxel being considered is ambiguous, choose the connections which result in smoothest surface. This algorithm works like wave propagation. Visualize the resulting mesh.

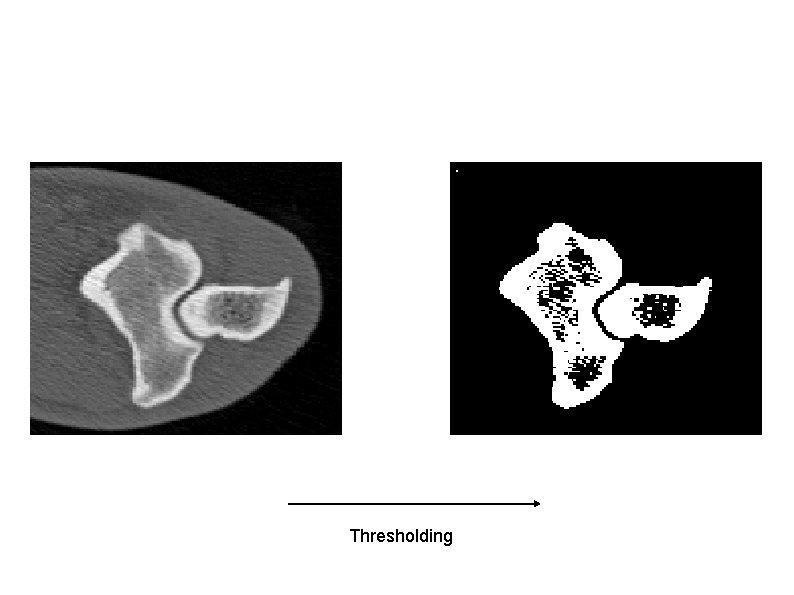
3 D Signed Distance Transform Requires a 3 D binary volume where 1's correspond to boundaries and 0's correspond to elsewhere. But, we have a gray level volume! We can obtain boundaries using segmentation. The output of simple thresholding + mathematical morphology is good enough.

3 D Signed Distance Transform Matlab implementation (bwdist) → very fast Ricardo Fabbri's implementation of 3 D distance transform explained in the paper: J. C. Torelli, R. Fabbri, G. Travieso, O. M. Bruno "A high performance 3 D exact Euclidean distance transform algorithm for distributed computing", International Journal of Pattern Recognition and Artificial Intelligence, 2010. It also requires boundary images (filled region images are not OK, because it treats every foreground pixel as boundary. ) So, I decided to continue with the matlab implementation.

Thresholding

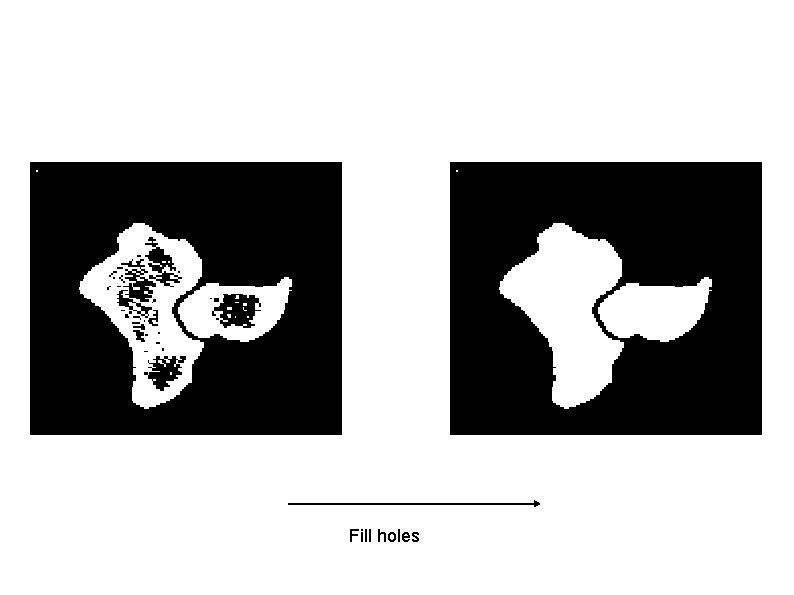
Fill holes

Median filtering

Erosion

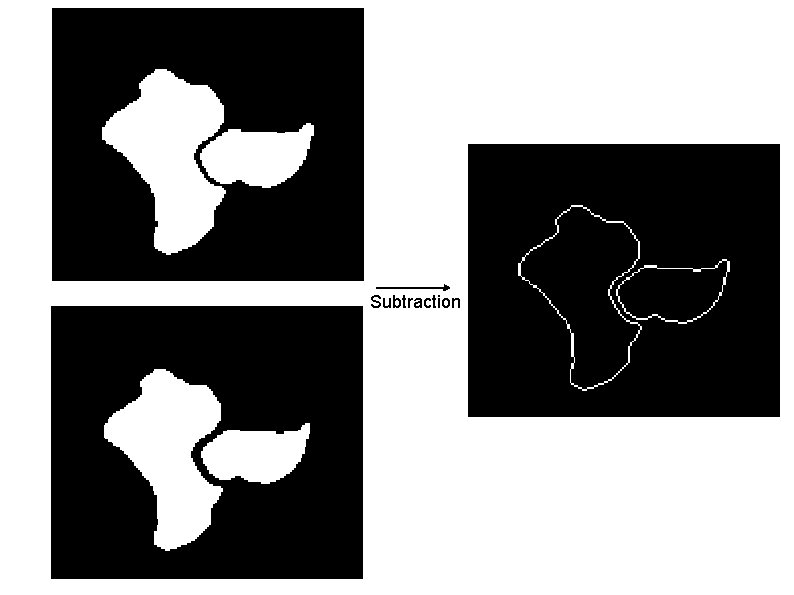
Subtraction

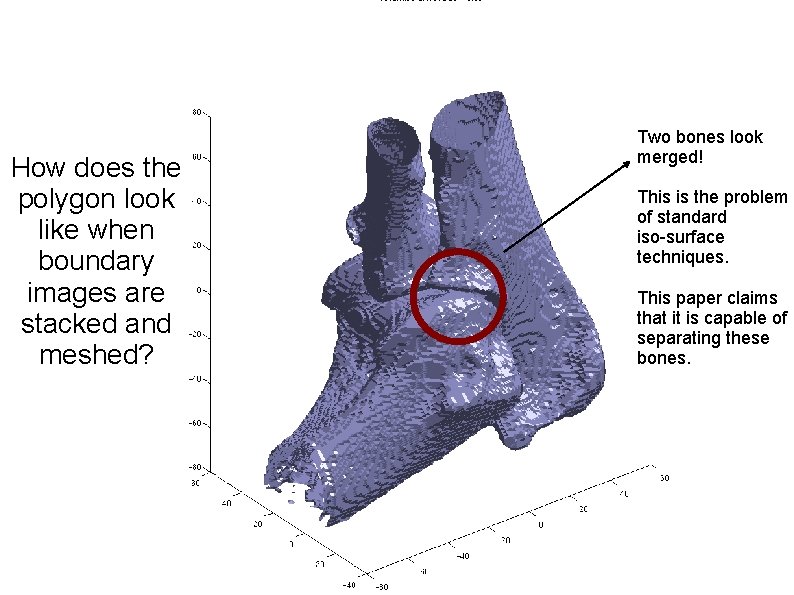
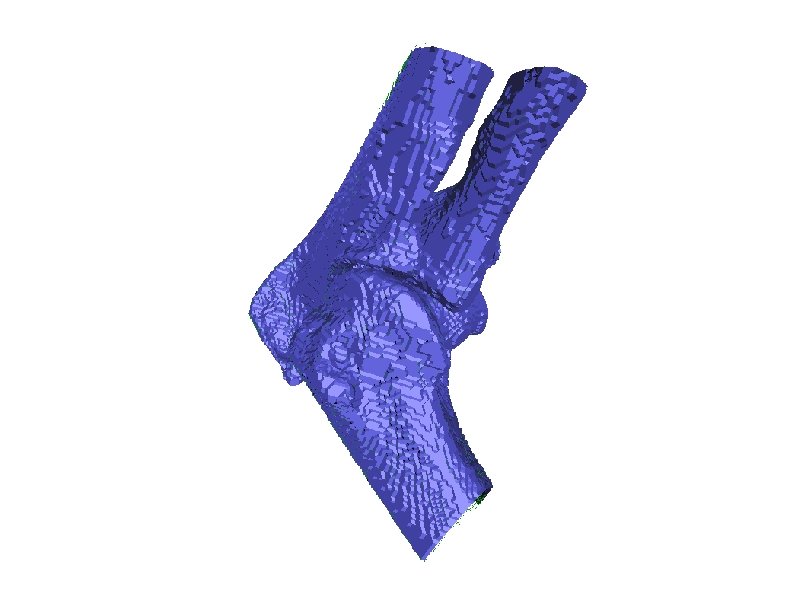
How does the polygon look like when boundary images are stacked and meshed? Two bones look merged! This is the problem of standard iso-surface techniques. This paper claims that it is capable of separating these bones.

3 D Signed Distance Transform Apply the operations to every slice → stack of binary boundary images Use matlab's bwdist function to obtain an unsigned distance transform. Multiply the interior regions by -1 to obtain a signed distance transform.

3 D Level Set Segmentation I might have used 3 D Level Set segmentation instead of binary segmentation + 3 D distance transform. VXL Implementations: Not clear which one to use? How to use? Found a matlab implementation Y. Zhang, B. J. Matuszewski, L. -K. Shark, and C. J. Moore. Medical Image Segmentation Using New Hybrid Level-Set Method. IEEE International Conference on Biomedical Visualisation, MEDi 08 VIS, London, pp. 7176, July, 2008. Takes a gray level volume and produces a distance transform.

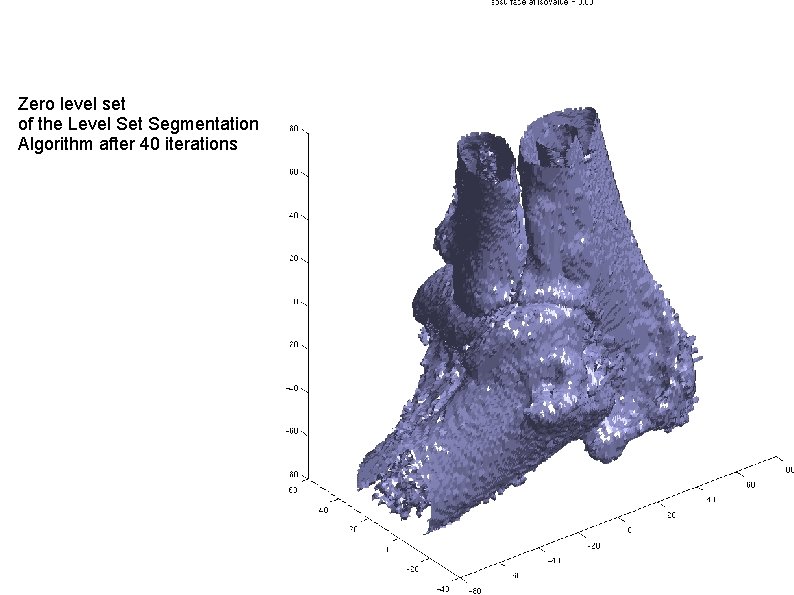
Zero level set of the Level Set Segmentation Algorithm after 40 iterations

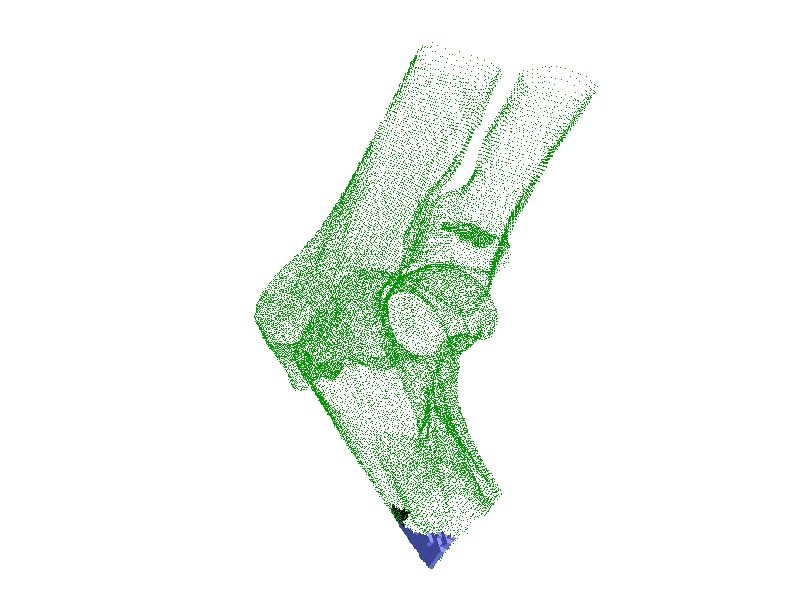
ENO Anchor Points “Subvoxel Polygonization of Discrete Implicit Surfaces Using ENO Interpolation” by Rodehorst and Kimia An old C/C++ implementation exists → not compatible with recent compilers Managed to revive the ENO anchor point extraction algorithm. Procedure: – Export the distance transform in a format that can be read by the ENO interpolation program. – Run ENO anchor point extraction code Input: 3 D signed distance transform For each grid line in each direction (x, y, z) Associate a 2 nd order polynomial with each interval between consecutive grid points using ENO interpolation. Output: Roots of the polynomials are the anchor points.

Wave Propagation Managed to revive the wave propagation component. Spent most of my time on this. Unfortunately, due to some bugs I couldn't resolve, it doesn't work on every input I provide. Doesn't work on 3 D level set output (finds odd number of ENO points and terminates). Works on binary segmentation + distance transform output.


Wave Propagation Example





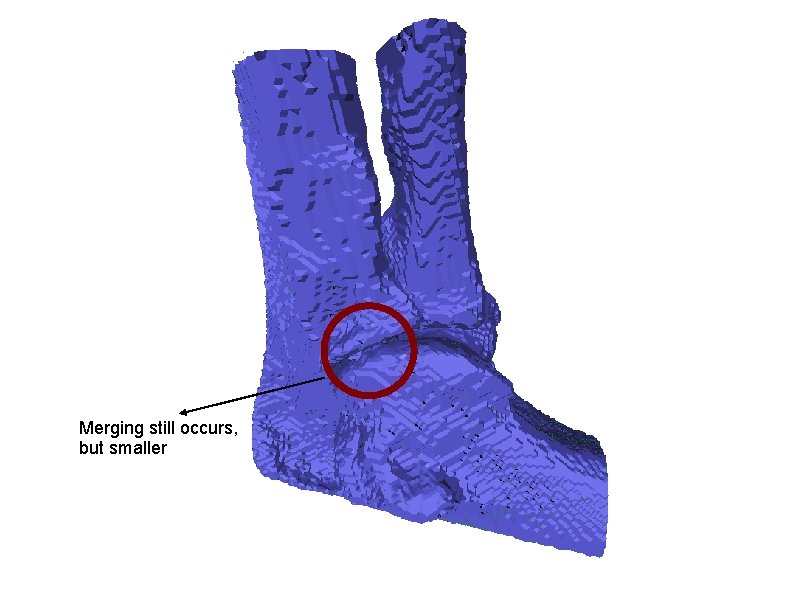
Merging still occurs, but smaller

More examples

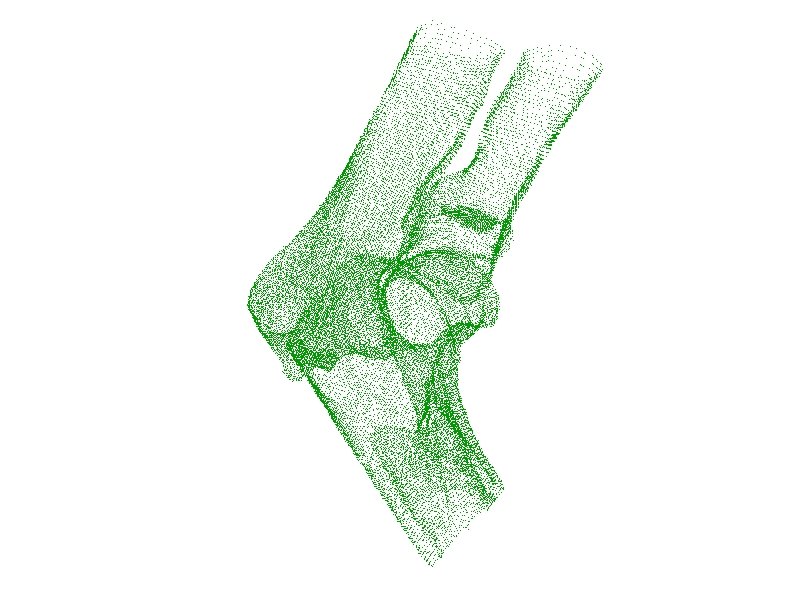
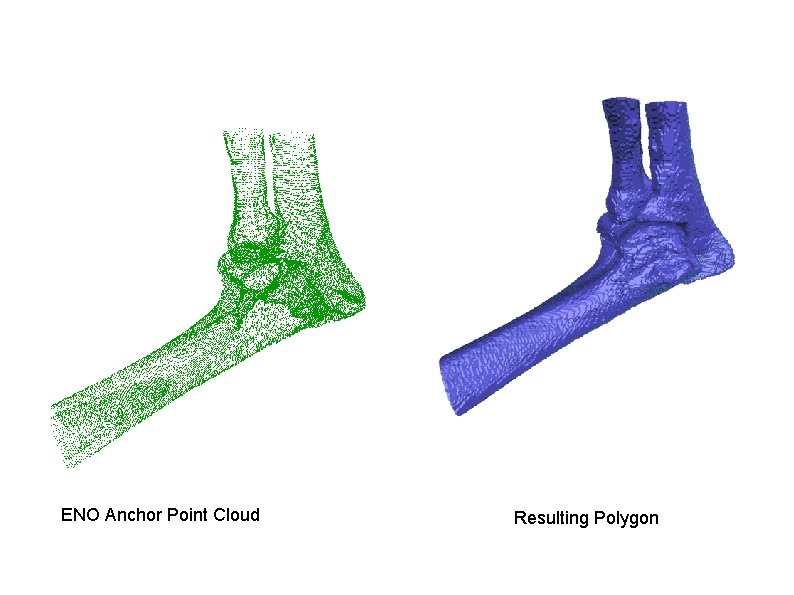
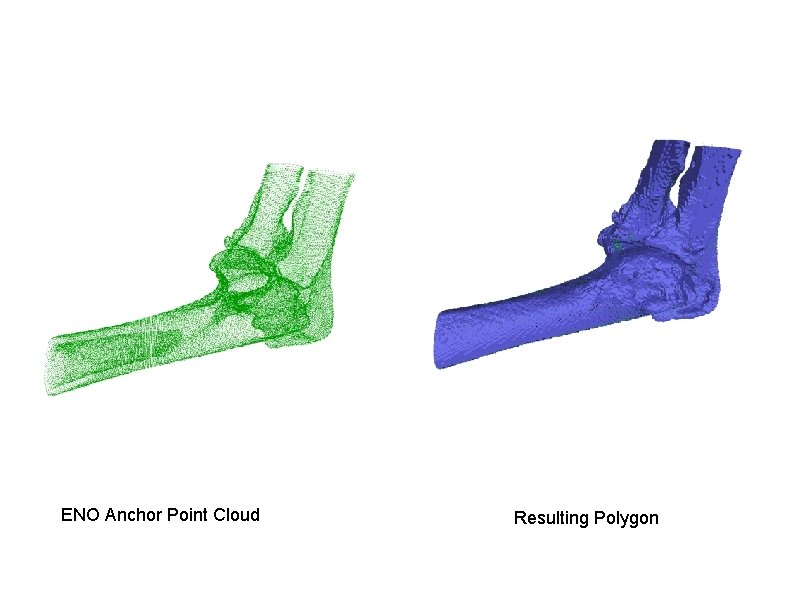
ENO Anchor Point Cloud Resulting Polygon

ENO Anchor Point Cloud Resulting Polygon

Conclusion Managed to revive the whole program Couldn't fix all the problems. The program still complains when level set segmentation results are used. Managed to reconstruct bones from 3 D CT volumes although the results still looked merged. Promising!