Engineering yeasts for next generation ethanol production Riaan





- Slides: 27

Engineering yeasts for next generation ethanol production Riaan den Haan 1, D. C. la Grange 1, M. Mert 1, H. Kroukamp 1, M. Saayman 1, M. Viktor 1, J. E. Mc. Bride 3, L. R. Lynd 3, M. Ilmen 4, M. Penttilä 4, J. F. Görgens 2, M. Bloom 1, W. H. van Zyl 1 (1) Depts. of Microbiology, and (2) Process Engineering Stellenbosch University, South Africa (3) Mascoma Corporation, Lebanon, NH (4) VTT Technical Research Centre of Finland

Introduction • Biofuels such as ethanol have gained significant interest due to environmental concerns and issues such as energy security - resulting in the current first generation ethanol market • Most of the ethanol produced worldwide is produced from starch • The development of a yeast that converts raw starch to ethanol in one step (CBP) could yield significant cost reductions in 1 st generation bioethanol production from corn starch • 2 nd Generation bioethanol produced from lignocellulosic biomass has great benefits in terms of energy balance, food security, etc. • Organisms that hydrolyse the cellulose and hemicelluloses in biomass and produce a valuable product such as ethanol at a high rate and titre would significantly reduce the costs of current biomass conversion technologies 2

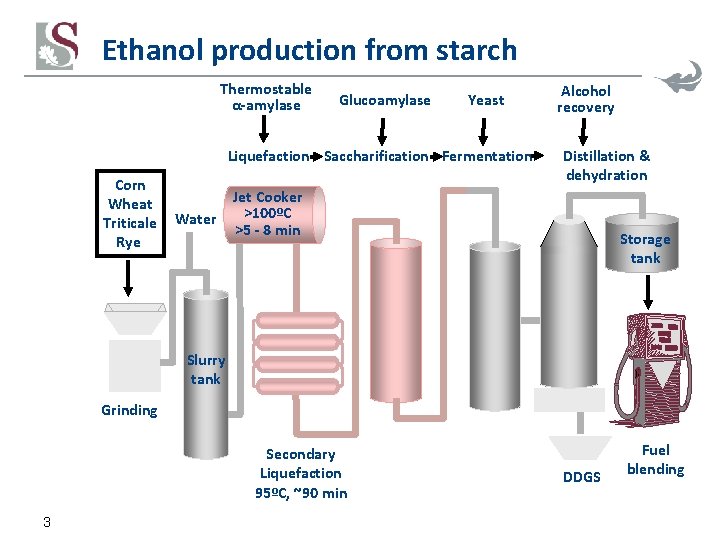
Ethanol production from starch Thermostable α-amylase Glucoamylase Yeast Liquefaction Saccharification Fermentation Corn Wheat Triticale Rye Water Alcohol recovery Distillation & dehydration Jet Cooker >100ºC >5 - 8 min Storage tank Slurry tank Grinding Secondary Liquefaction 95ºC, ~90 min 3 DDGS Fuel blending

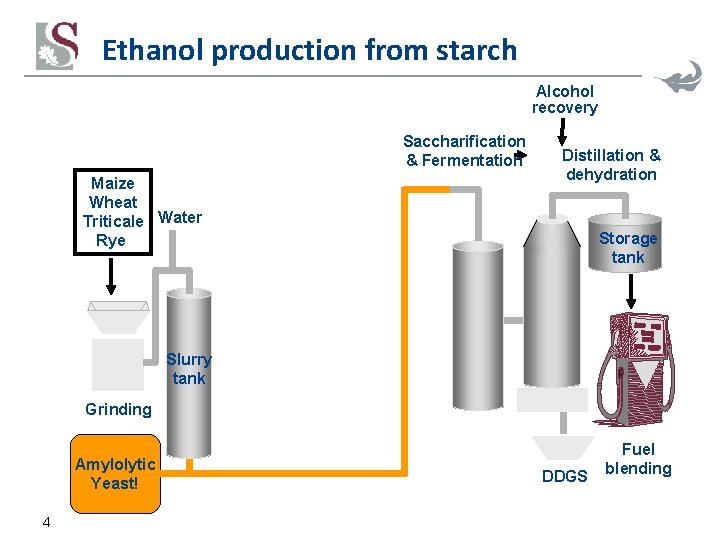
Ethanol production from starch Alcohol recovery Saccharification & Fermentation Maize Wheat Triticale Water Rye Distillation & dehydration Storage tank Slurry tank Grinding Amylolytic Yeast! 4 DDGS Fuel blending

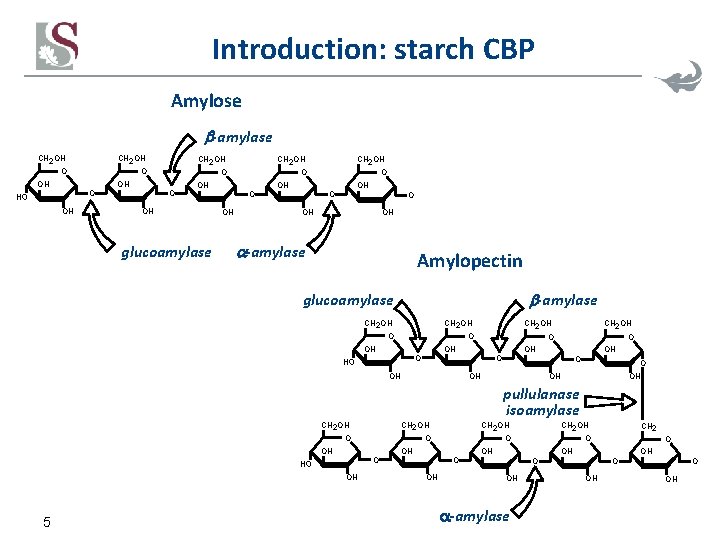
Introduction: starch CBP Amylose -amylase CH 2 OH O OH CH 2 OH O O OH OH CH 2 OH OH glucoamylase CH 2 OH O O OH OH -amylase Amylopectin -amylase glucoamylase CH 2 OH O OH OH CH 2 OH O OH OH pullulanase isoamylase CH 2 OH O CH 2 OH OH 5 O OH -amylase O O OH

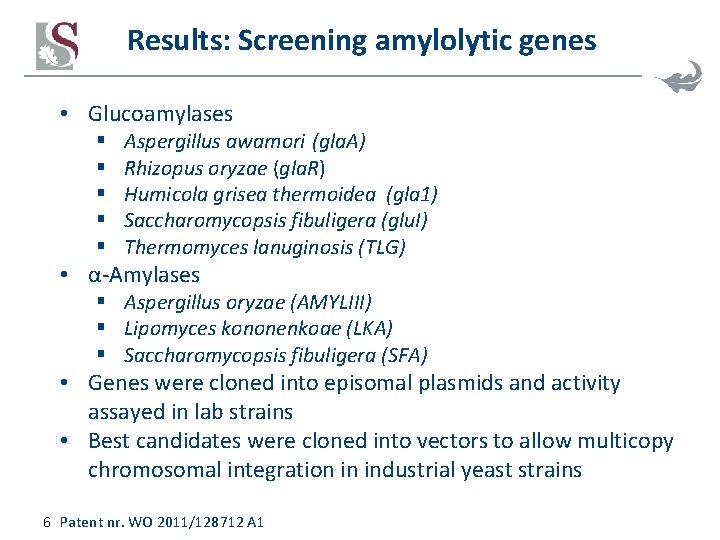
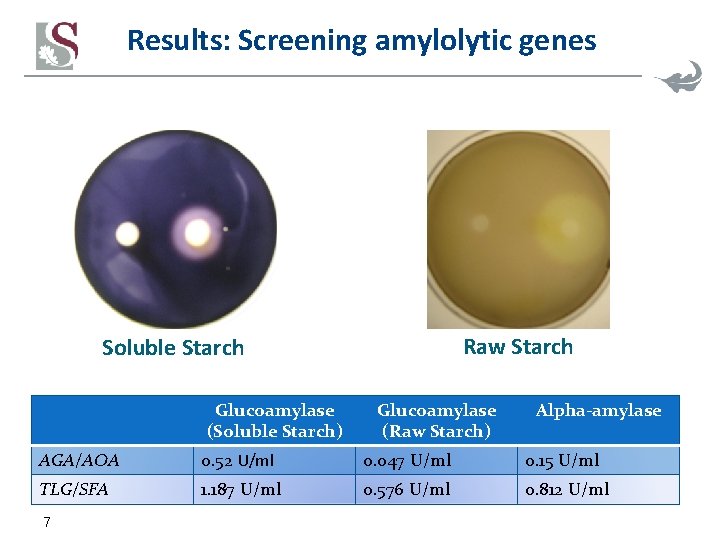
Results: Screening amylolytic genes • Glucoamylases § § § Aspergillus awamori (gla. A) Rhizopus oryzae (gla. R) Humicola grisea thermoidea (gla 1) Saccharomycopsis fibuligera (glu. I) Thermomyces lanuginosis (TLG) • α-Amylases § Aspergillus oryzae (AMYLIII) § Lipomyces kononenkoae (LKA) § Saccharomycopsis fibuligera (SFA) • Genes were cloned into episomal plasmids and activity assayed in lab strains • Best candidates were cloned into vectors to allow multicopy chromosomal integration in industrial yeast strains 6 Patent nr. WO 2011/128712 A 1

Results: Screening amylolytic genes Raw Starch Soluble Starch Glucoamylase (Soluble Starch) Glucoamylase (Raw Starch) Alpha-amylase AGA/AOA 0. 52 U/ml 0. 047 U/ml 0. 15 U/ml TLG/SFA 1. 187 U/ml 0. 576 U/ml 0. 812 U/ml 7

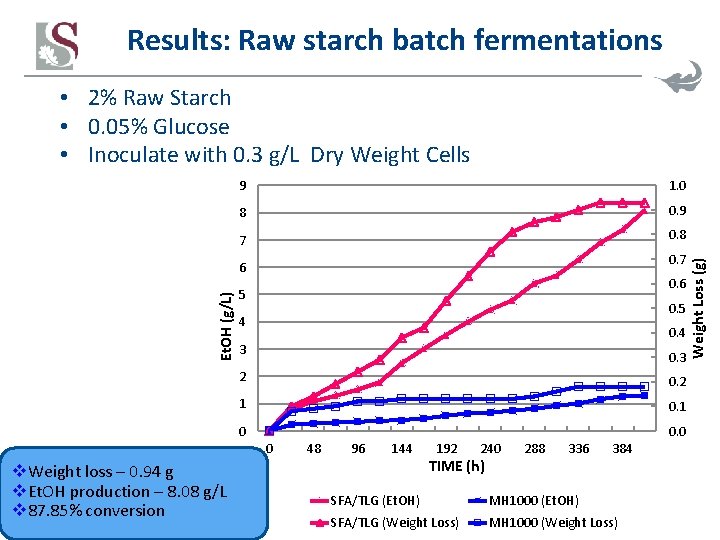
Results: Raw starch batch fermentations 9 1. 0 8 0. 9 7 0. 8 0. 7 Et. OH (g/L) 6 0. 6 5 0. 5 4 0. 4 3 0. 3 2 0. 2 1 0 0. 0 0 v. Weight loss – 0. 94 g v. Et. OH production – 8. 08 g/L v 87. 85% conversion 48 96 144 192 240 TIME (h) 288 336 384 SFA/TLG (Et. OH) MH 1000 (Et. OH) SFA/TLG (Weight Loss) MH 1000 (Weight Loss) Weight Loss (g) • 2% Raw Starch • 0. 05% Glucose • Inoculate with 0. 3 g/L Dry Weight Cells

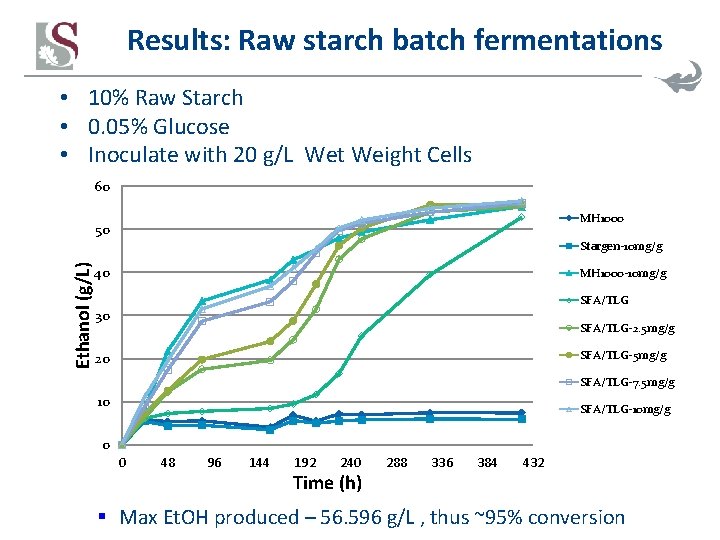
Results: Raw starch batch fermentations • 10% Raw Starch • 0. 05% Glucose • Inoculate with 20 g/L Wet Weight Cells 60 MH 1000 50 Ethanol (g/L) Stargen-10 mg/g 40 MH 1000 -10 mg/g SFA/TLG 30 SFA/TLG-2. 5 mg/g SFA/TLG-5 mg/g 20 SFA/TLG-7. 5 mg/g 10 SFA/TLG-10 mg/g 0 0 48 96 144 192 240 Time (h) 288 336 384 432 § Max Et. OH produced – 56. 596 g/L , thus ~95% conversion

Discussion: Starch CBP • Raw starch conversion was possible with no added enzymes or with reduced enzyme loadings; fermentation times must be improved • Current and future prospects: • • • 10 Screen yeast strains with superior fermentation capacities Screen a wider array of α-amylase encoding genes Create strain with higher copy numbers of genes

Introduction: Lignocellulose CBP • Lignocellulosic biomass consisting of mainly lignin, cellulose and hemicellulose, is an abundant, renewable & sustainable source of fuels etc. • The main barrier that prevents widespread utilization of this resource for production of commodity products is the lack of low-cost technologies to overcome the recalcitrance of lignocellulose 11

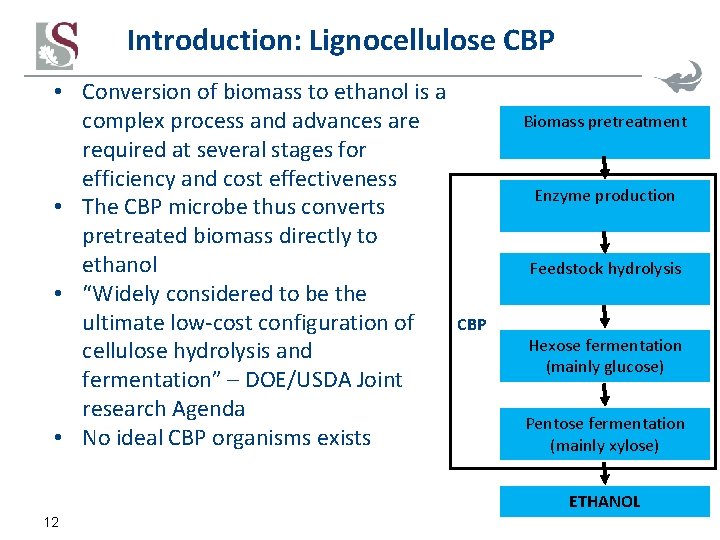
Introduction: Lignocellulose CBP • Conversion of biomass to ethanol is a complex process and advances are required at several stages for efficiency and cost effectiveness • The CBP microbe thus converts pretreated biomass directly to ethanol • “Widely considered to be the ultimate low-cost configuration of CBP cellulose hydrolysis and fermentation” – DOE/USDA Joint research Agenda • No ideal CBP organisms exists Biomass pretreatment Enzyme production Feedstock hydrolysis Hexose fermentation (mainly glucose) Pentose fermentation (mainly xylose) ETHANOL 12

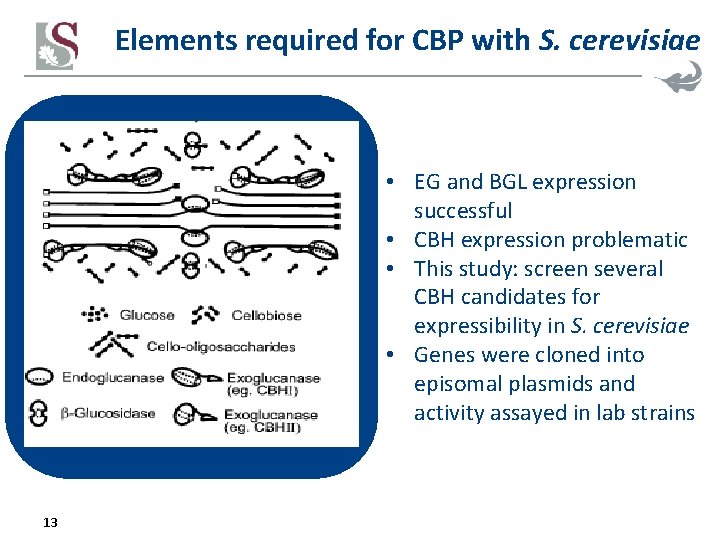
Elements required for CBP with S. cerevisiae • EG and BGL expression successful • CBH expression problematic • This study: screen several CBH candidates for expressibility in S. cerevisiae • Genes were cloned into episomal plasmids and activity assayed in lab strains 13

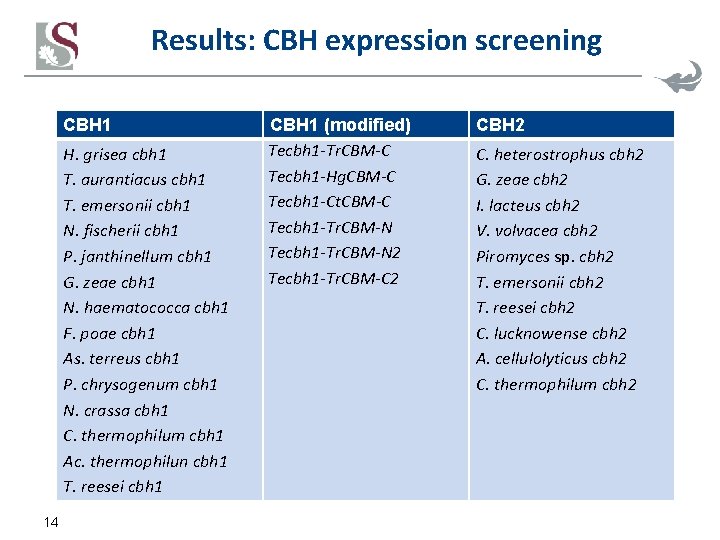
Results: CBH expression screening CBH 1 H. grisea cbh 1 T. aurantiacus cbh 1 T. emersonii cbh 1 N. fischerii cbh 1 P. janthinellum cbh 1 G. zeae cbh 1 N. haematococca cbh 1 F. poae cbh 1 As. terreus cbh 1 P. chrysogenum cbh 1 N. crassa cbh 1 C. thermophilum cbh 1 Ac. thermophilun cbh 1 T. reesei cbh 1 14 CBH 1 (modified) Tecbh 1 -Tr. CBM-C Tecbh 1 -Hg. CBM-C Tecbh 1 -Ct. CBM-C Tecbh 1 -Tr. CBM-N 2 Tecbh 1 -Tr. CBM-C 2 CBH 2 C. heterostrophus cbh 2 G. zeae cbh 2 I. lacteus cbh 2 V. volvacea cbh 2 Piromyces sp. cbh 2 T. emersonii cbh 2 T. reesei cbh 2 C. lucknowense cbh 2 A. cellulolyticus cbh 2 C. thermophilum cbh 2

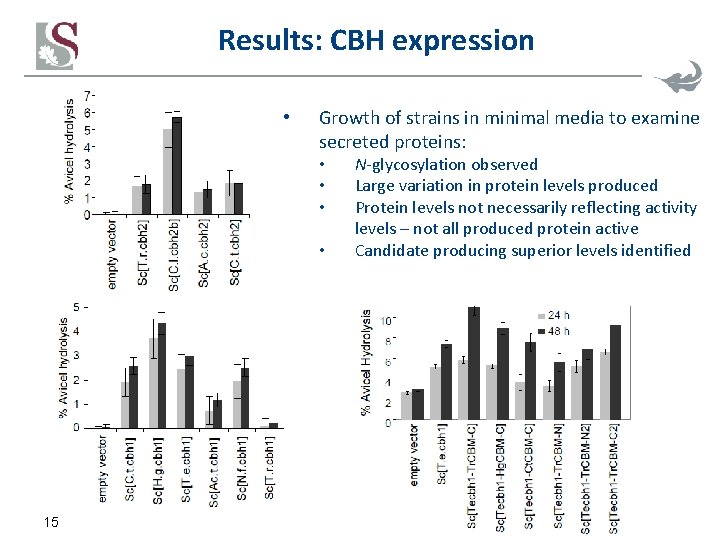
Results: CBH expression • Growth of strains in minimal media to examine secreted proteins: • • 15 N-glycosylation observed Large variation in protein levels produced Protein levels not necessarily reflecting activity levels – not all produced protein active Candidate producing superior levels identified

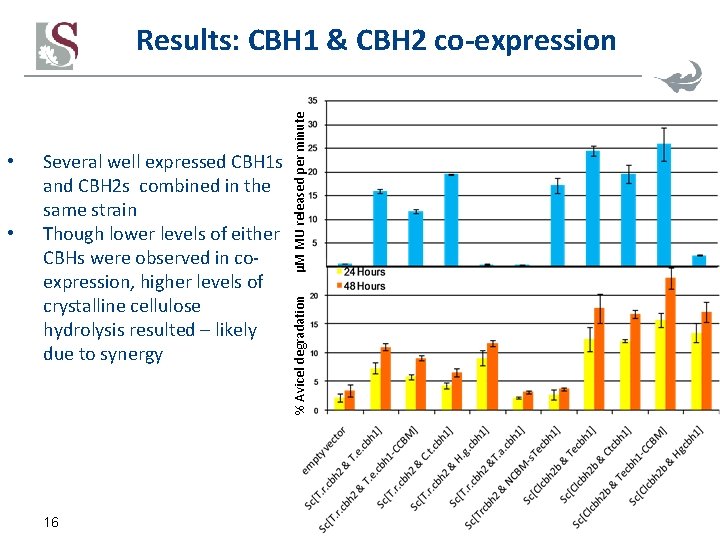
• • Several well expressed CBH 1 s and CBH 2 s combined in the same strain Though lower levels of either CBHs were observed in coexpression, higher levels of crystalline cellulose hydrolysis resulted – likely due to synergy 16 % Avicel degradation μM MU released per minute Results: CBH 1 & CBH 2 co-expression

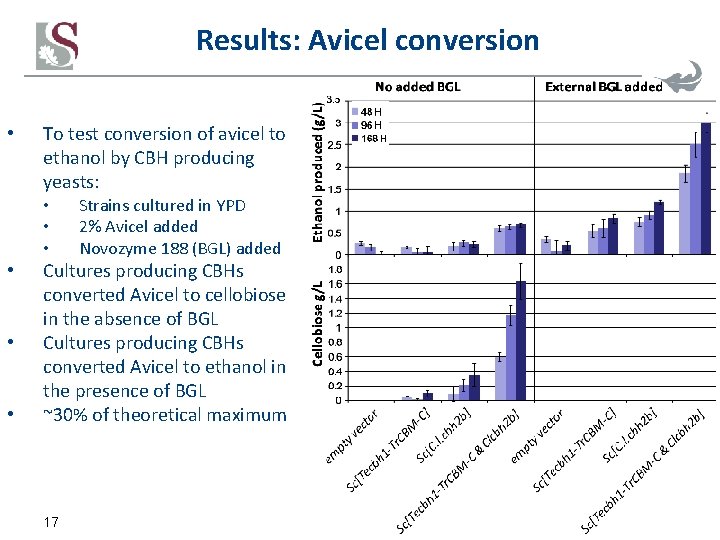
Results: Avicel conversion • To test conversion of avicel to ethanol by CBH producing yeasts: • • • Strains cultured in YPD 2% Avicel added Novozyme 188 (BGL) added Cultures producing CBHs converted Avicel to cellobiose in the absence of BGL Cultures producing CBHs converted Avicel to ethanol in the presence of BGL ~30% of theoretical maximum 17

Discussion: cellulose CBP • • • 18 High level secretion of exoglucanases is required for crystalline cellulose utilization - major hurdle in CBP yeast development Indentified gene candidates compatible with expression in yeast § T. e. CBH 1 and its T. r. CBM attached derivative yielded 100 -200 mg/liter in shake flasks and ~300 mg/liter in HCD conditions § The highest CBH level secreted, ~1 g/liter C. l. CBH 2 b (~4% tcp) exceeded any previous reports on CBH production in S. cerevisiae Thus S. cerevisiae is capable of secreting CBHs at high levels that compare well with the highest heterologous protein production levels described for S. cerevisiae

Introduction: strain engineering • • • 19 The innate low secretion capacity of S. cerevisiae, even when compared to other yeast species represents a drawback in its development as a CBP organism Over-expression of genes encoding foldases, chaperones or other parts of the secretion pathways or knockouts of genes encoding negative regulators have been shown to increase secretion capacity in fungi We aimed to improve the secretion of hydrolases by S. cerevisiae through strain engineering

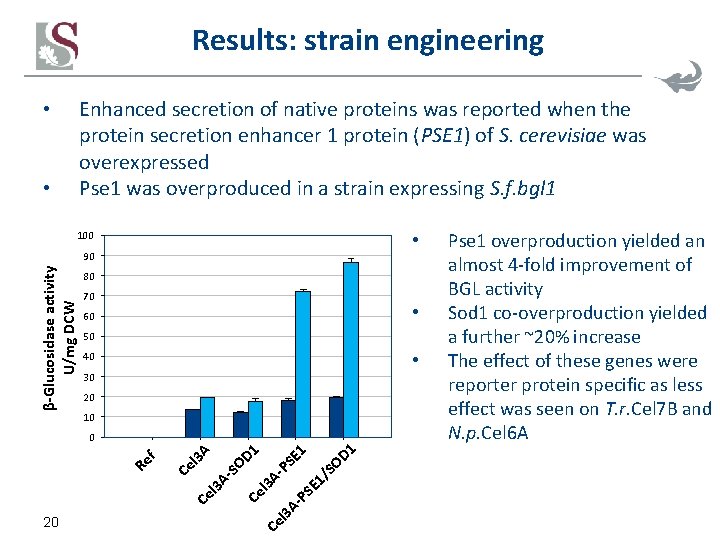
Results: strain engineering Enhanced secretion of native proteins was reported when the protein secretion enhancer 1 protein (PSE 1) of S. cerevisiae was overexpressed Pse 1 was overproduced in a strain expressing S. f. bgl 1 • • • 100 β-Glucosidase activity U/mg DCW 90 80 70 • 60 50 • 40 30 20 10 l 3 A- PS E 1 / SO D 1 E 1 PS A- D 1 Ce 20 Ce l 3 A- SO A Ce l 3 Re f 0 Pse 1 overproduction yielded an almost 4 -fold improvement of BGL activity Sod 1 co-overproduction yielded a further ~20% increase The effect of these genes were reporter protein specific as less effect was seen on T. r. Cel 7 B and N. p. Cel 6 A

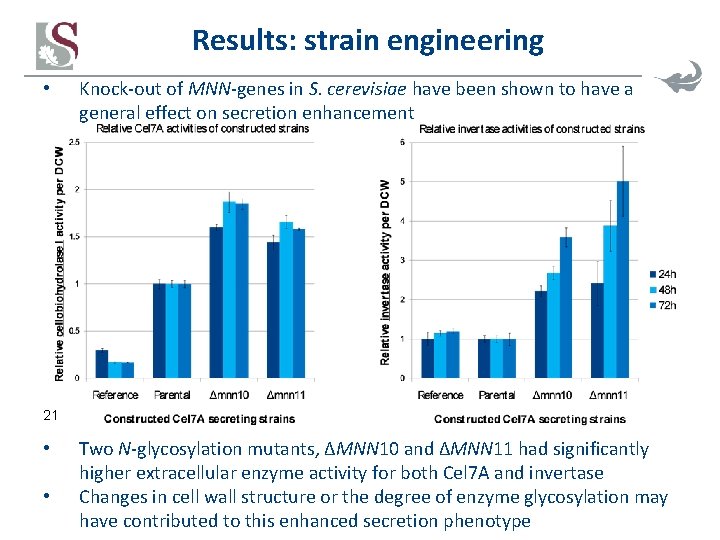
Results: strain engineering • Knock-out of MNN-genes in S. cerevisiae have been shown to have a general effect on secretion enhancement 21 • • Two N-glycosylation mutants, ΔMNN 10 and ΔMNN 11 had significantly higher extracellular enzyme activity for both Cel 7 A and invertase Changes in cell wall structure or the degree of enzyme glycosylation may have contributed to this enhanced secretion phenotype

Conclusion • • 22 Fermentation of raw starch by recombinant S. cerevisiae strains was demonstrated without the addition of commercial enzymes S. cerevisiae was shown to be capable of expression of levels of CBHs that would overcome the barrier of sufficiency for conversion of cellulosic biomass to ethanol § Simultaneous expression of CBHs with EG and β-glucosidase enabled S. cerevisiae to directly convert cellulosic substrates to ethanol and to grow on cellulose under CBP conditions S. cerevisiae strains could be manipulated to allow improved secretion of hydrolase enzymes Combining optimal gene candidates in enhanced host strains will lead to improved strains for CBP applications

Thank you! Acknowledgments: Lee Lynd John Mc. Bride Elena Brevnova Allan Froehlich Alan Gilbert Heidi Hau Erin Wiswall Hoowen Xu Merja Penttilä Marja Ilmen Anu Koivula Sanni Voutilainen 23 Emile van Zyl Riaan den Haan Marlin Mert Danie La Grange Maryna Saayman Marko Viktor Heinrich Kroukamp

Barriers to lignocellulose CBP with S. cerevisiae 1. Consumption of all major sugar constituents of biomass 2. High level expression of cellulases, especially cellobiohydrolases 3. Expression of the diverse enzymes required to hydrolyze biomass 4. Production of enzymes and consumption of sugars in toxic process conditions 24

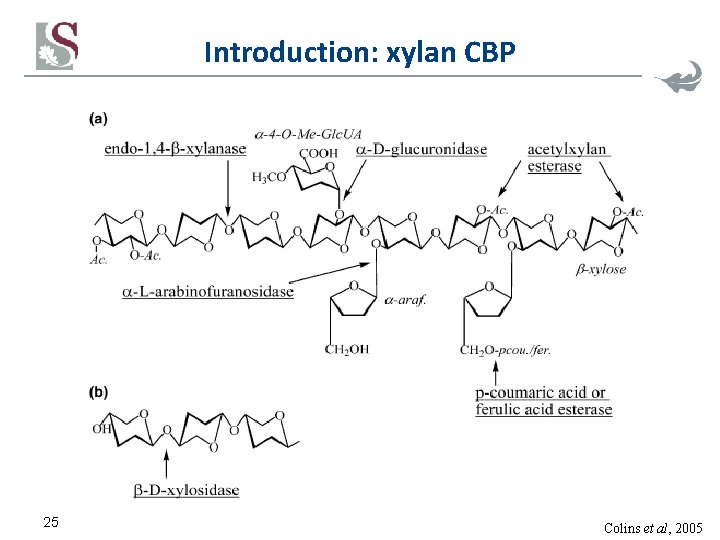
Introduction: xylan CBP 25 Colins et al, 2005

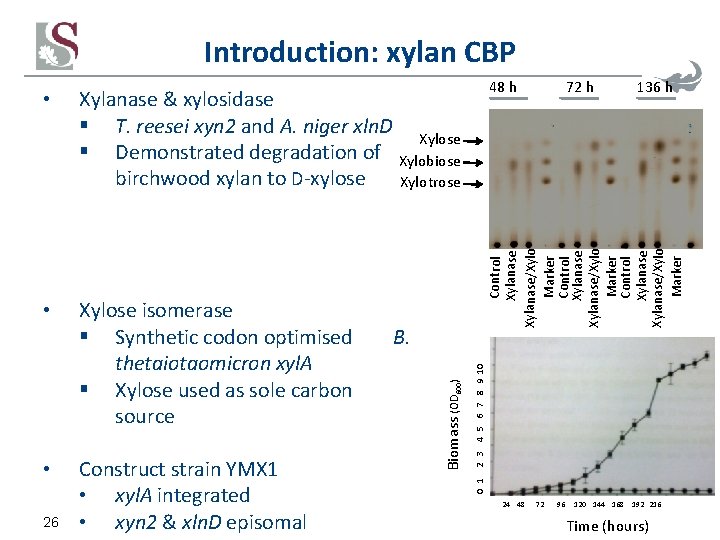
Introduction: xylan CBP Xylanase & xylosidase § T. reesei xyn 2 and A. niger xln. D Xylose § Demonstrated degradation of Xylobiose birchwood xylan to D-xylose Xylotrose • Xylose isomerase § Synthetic codon optimised B. thetaiotaomicron xyl. A § Xylose used as sole carbon source • Construct strain YMX 1 • xyl. A integrated • xyn 2 & xln. D episomal 26 72 h 136 h 0 1 2 3 4 5 6 7 8 9 10 Biomass (OD 600) Control Xylanase/Xylo Marker Control Xylanase/Xylo Marker • 48 h 24 48 72 96 120 144 168 192 216 Time (hours)

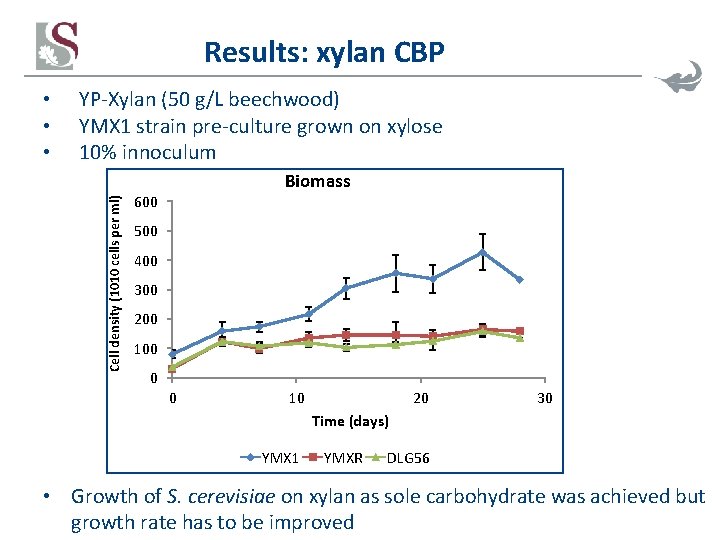
Results: xylan CBP YP-Xylan (50 g/L beechwood) YMX 1 strain pre-culture grown on xylose 10% innoculum Cell density (1010 cells per ml) • • • Biomass 600 500 400 300 200 100 0 0 10 20 30 Time (days) YMX 1 YMXR DLG 56 • Growth of S. cerevisiae on xylan as sole carbohydrate was achieved but growth rate has to be improved