ENCODE The ENCODE Project Consortium EBI is an

ENCODE The ENCODE Project Consortium EBI is an Outstation of the European Molecular Biology Laboratory.
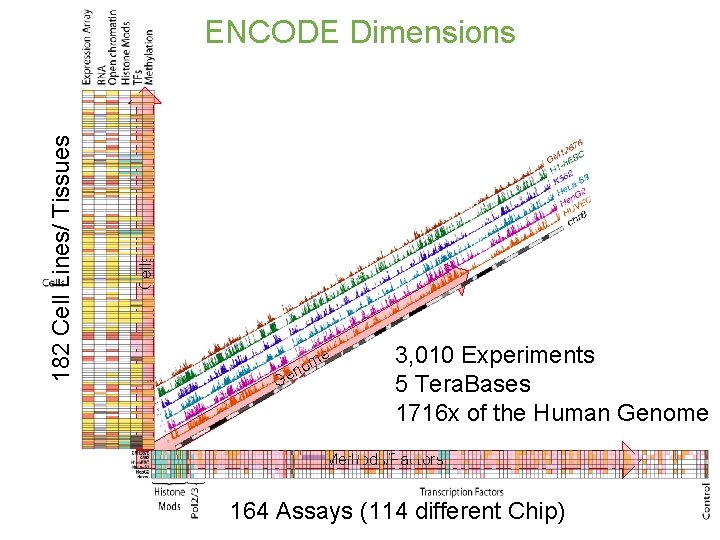
Cells 182 Cell Lines/ Tissues ENCODE Dimensions Ge m no e 3, 010 Experiments 5 Tera. Bases 1716 x of the Human Genome Methods/Factors 164 Assays (114 different Chip)

Basic Processing Anshul Kundaje, Qun’hua Li. Steve Wilder, Ben Brown, Joel Rozowsky, Felix Schlesinger, Arif Ozgun Harmanci EBI is an Outstation of the European Molecular Biology Laboratory.
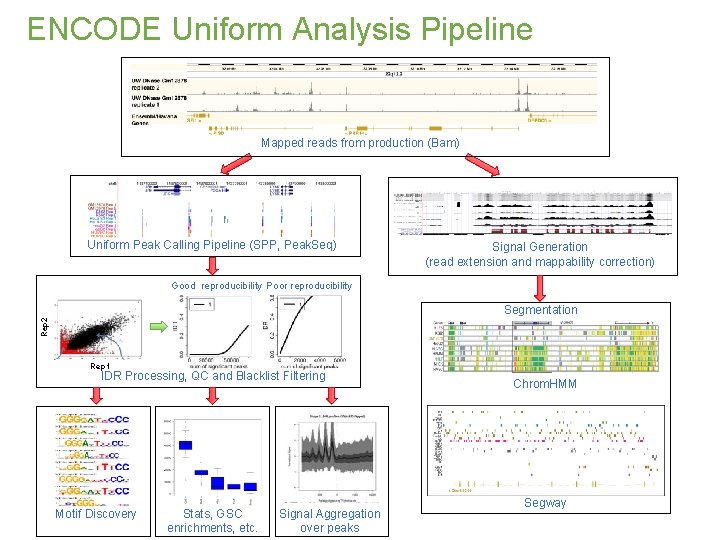
ENCODE Uniform Analysis Pipeline Mapped reads from production (Bam) Uniform Peak Calling Pipeline (SPP, Peak. Seq) Signal Generation (read extension and mappability correction) Good reproducibility Poor reproducibility Rep 2 Segmentation Rep 1 IDR Processing, QC and Blacklist Filtering Motif Discovery Stats, GSC enrichments, etc. Signal Aggregation over peaks Chrom. HMM Segway
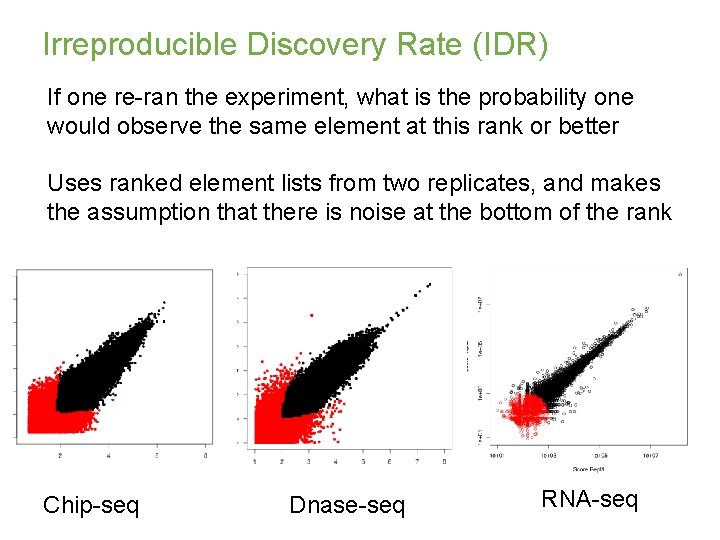
Irreproducible Discovery Rate (IDR) If one re-ran the experiment, what is the probability one would observe the same element at this rank or better Uses ranked element lists from two replicates, and makes the assumption that there is noise at the bottom of the rank Chip-seq Dnase-seq RNA-seq
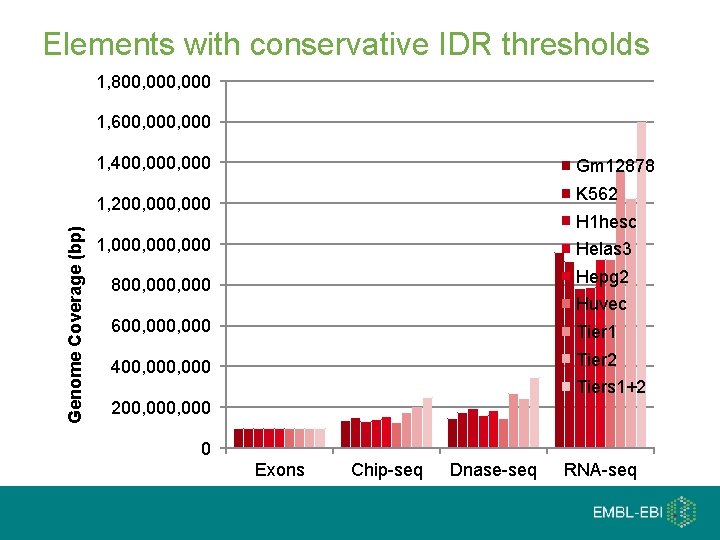
Elements with conservative IDR thresholds 1, 800, 000 1, 600, 000 1, 400, 000 Gm 12878 K 562 Genome Coverage (bp) 1, 200, 000 H 1 hesc 1, 000, 000 Helas 3 800, 000 Hepg 2 600, 000 Tier 1 400, 000 Tier 2 Huvec Tiers 1+2 200, 000 0 Exons Chip-seq Dnase-seq RNA-seq
Organisation+Access for the data wwww. encodeproject. org (UCSC) UCSC Genome Browser Ensembl “Factorbook”

Ongoing analyses in Genome Biology Javier Herrero, Xianjun Dong, Zhiping Weng, Mark Gerstein, Bob Altschuler, Tim Reddy, Chao Cheng, Koon-kiu Yan, Michael Hoffman, Jason Ernst, Anshul Kundaje, Chao Cheng, Manolis Kellis, Mark Gerstein, Kevin Yip, Bill Noble, Ali Mortazavi, Belinda Giardine EBI is an Outstation of the European Molecular Biology Laboratory.
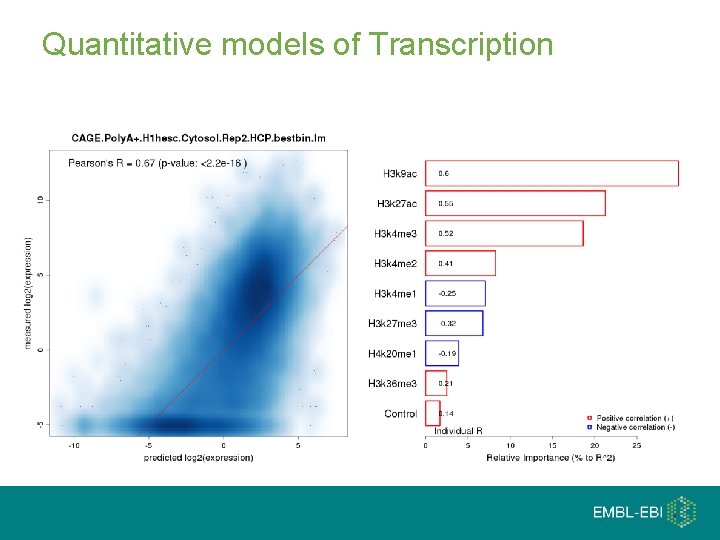
Quantitative models of Transcription
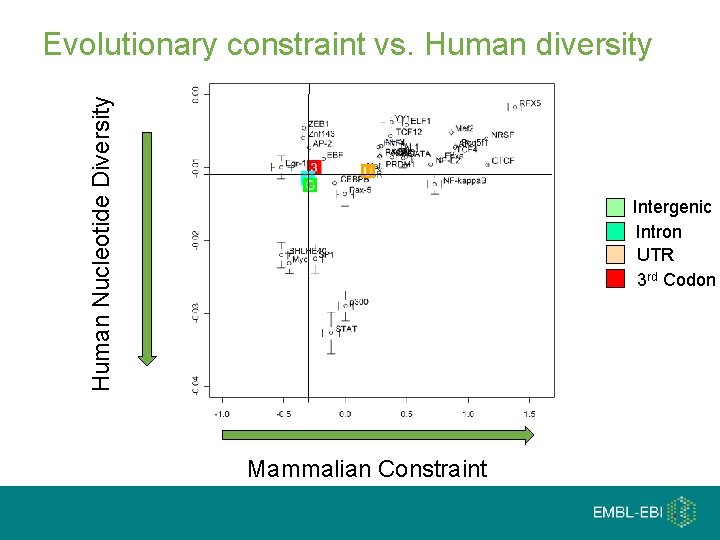
Human Nucleotide Diversity Evolutionary constraint vs. Human diversity Intergenic Intron UTR 3 rd Codon Mammalian Constraint
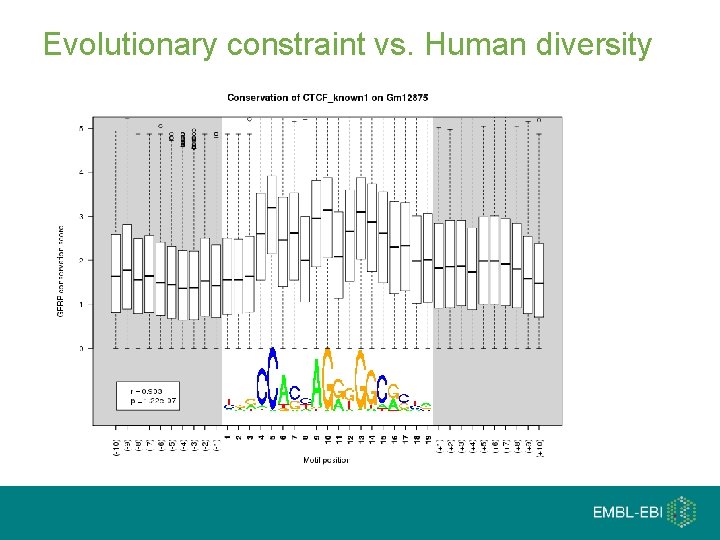
Evolutionary constraint vs. Human diversity
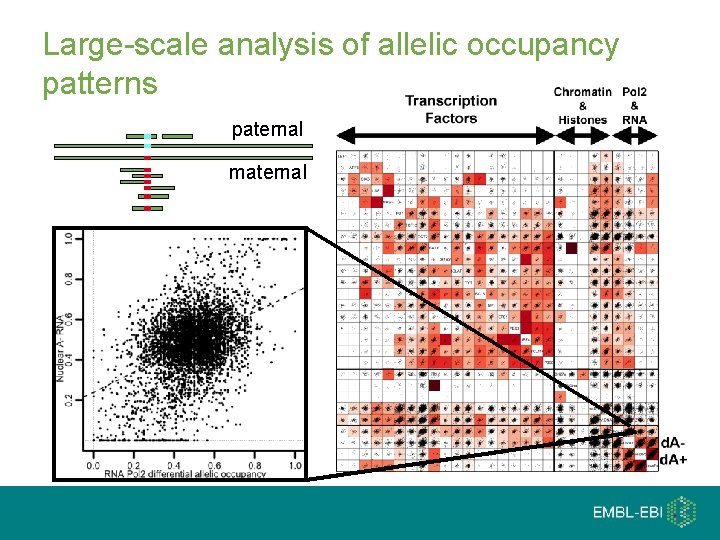
Large-scale analysis of allelic occupancy patterns paternal maternal
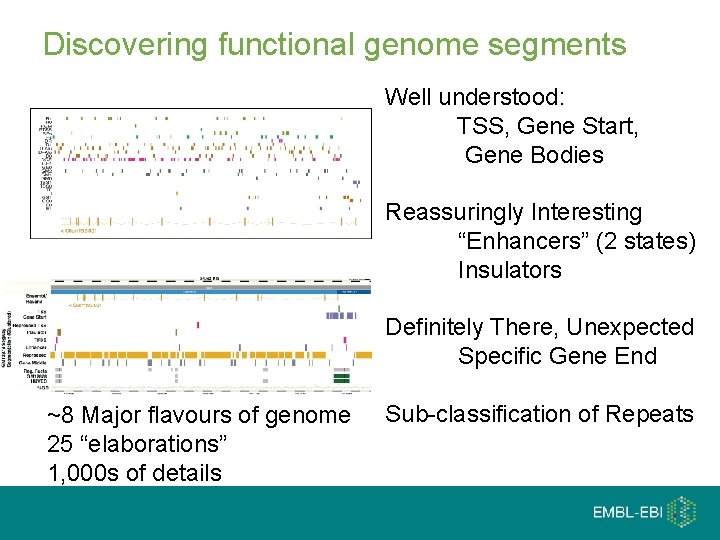
Discovering functional genome segments Well understood: TSS, Gene Start, Gene Bodies Reassuringly Interesting “Enhancers” (2 states) Insulators Definitely There, Unexpected Specific Gene End ~8 Major flavours of genome 25 “elaborations” 1, 000 s of details Sub-classification of Repeats
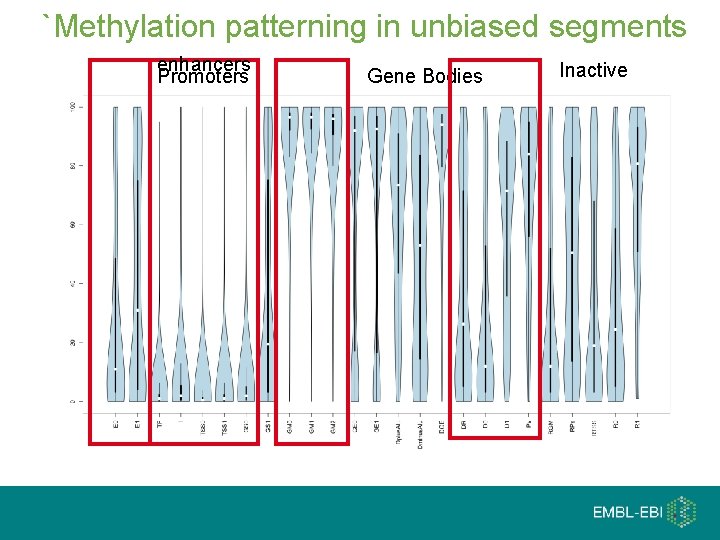
`Methylation patterning in unbiased segments enhancers Promoters Gene Bodies Inactive
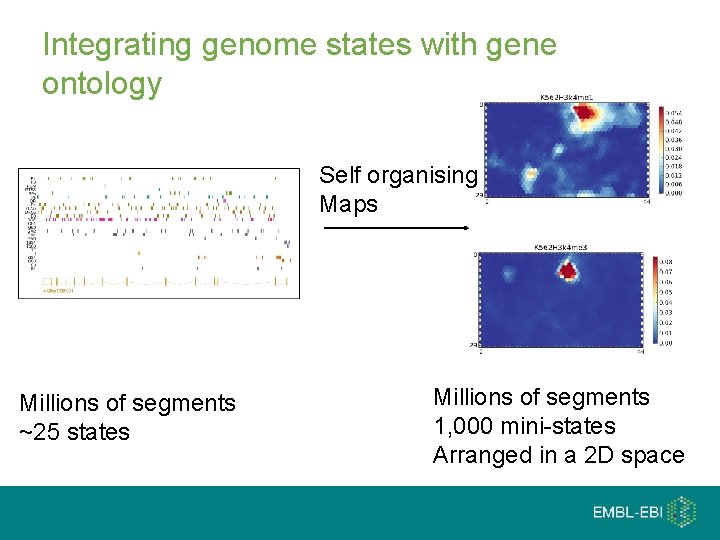
Integrating genome states with gene ontology Self organising Maps Millions of segments ~25 states Millions of segments 1, 000 mini-states Arranged in a 2 D space
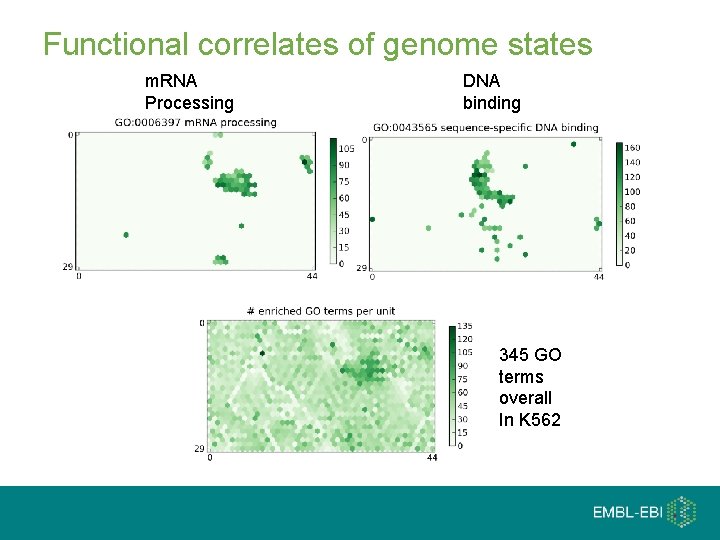
Functional correlates of genome states m. RNA Processing DNA binding 345 GO terms overall In K 562
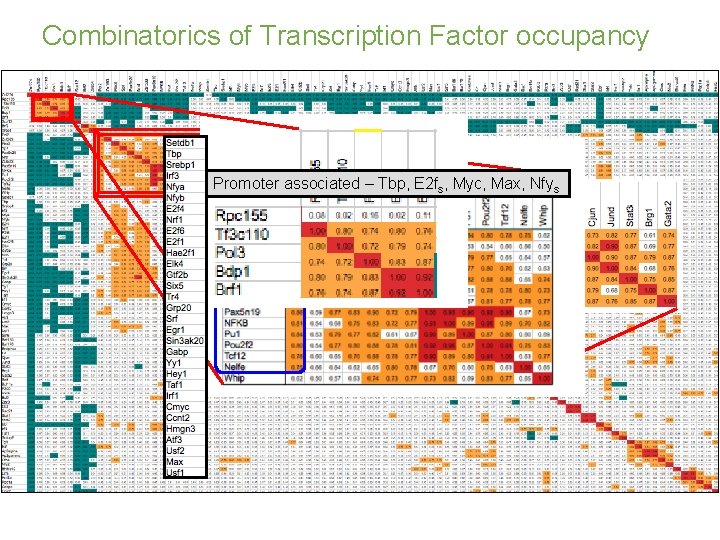
Combinatorics of Transcription Factor occupancy Promoter associated – Tbp, E 2 fs, Myc, Max, Nfys
Visualizing transcription factor connectivity

Impact on Understanding Variation Ross Hardison, Mark Gerstein, Alexej Abyzov, Xinmeng Jasmine Mu, John Stamatoyannopoulos, Joel Rozowsky EBI is an Outstation of the European Molecular Biology Laboratory.
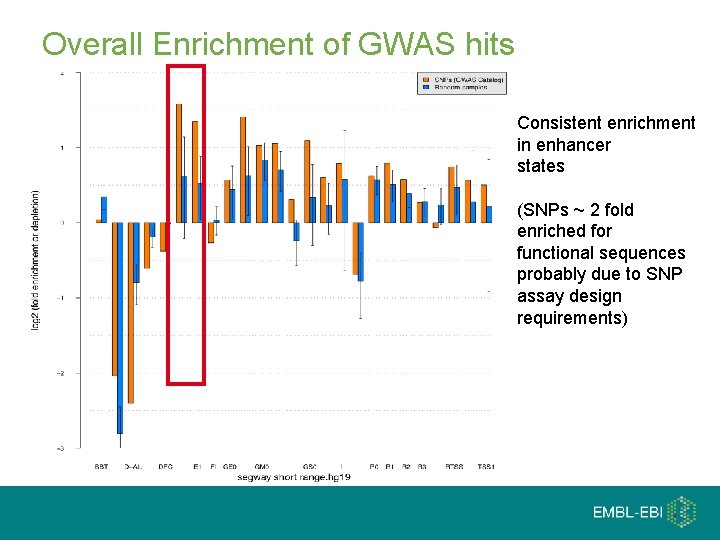
Overall Enrichment of GWAS hits Consistent enrichment in enhancer states (SNPs ~ 2 fold enriched for functional sequences probably due to SNP assay design requirements)
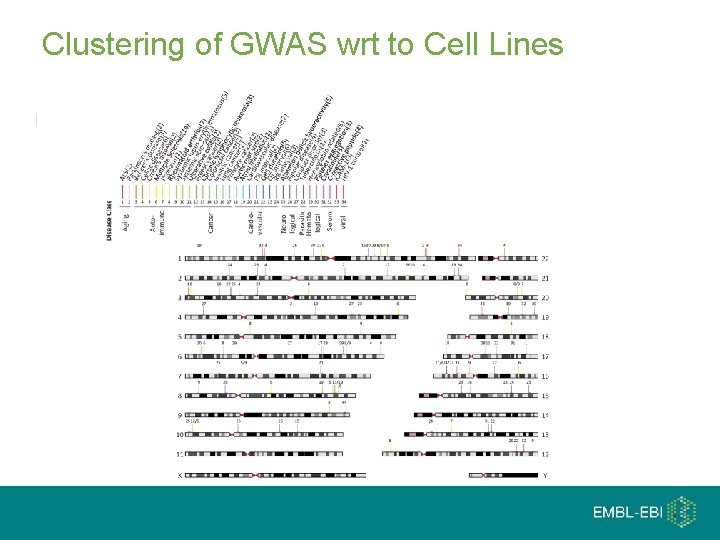
Clustering of GWAS wrt to Cell Lines
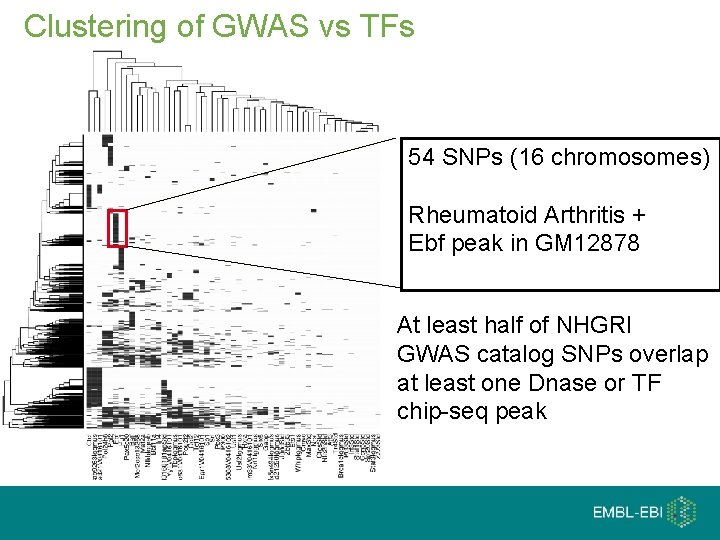
Clustering of GWAS vs TFs 54 SNPs (16 chromosomes) Rheumatoid Arthritis + Ebf peak in GM 12878 At least half of NHGRI GWAS catalog SNPs overlap at least one Dnase or TF chip-seq peak
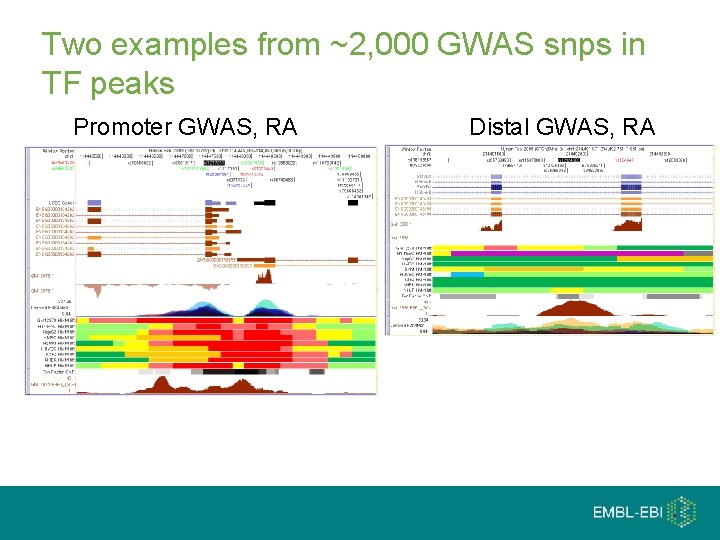
Two examples from ~2, 000 GWAS snps in TF peaks Promoter GWAS, RA Distal GWAS, RA
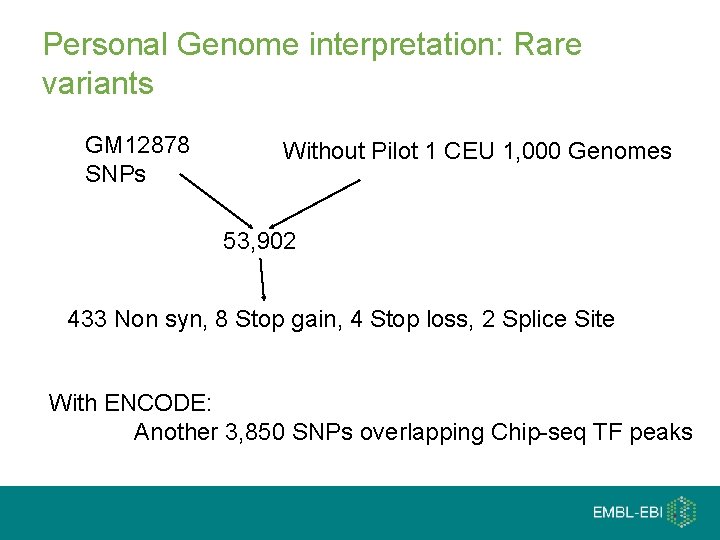
Personal Genome interpretation: Rare variants GM 12878 SNPs Without Pilot 1 CEU 1, 000 Genomes 53, 902 433 Non syn, 8 Stop gain, 4 Stop loss, 2 Splice Site With ENCODE: Another 3, 850 SNPs overlapping Chip-seq TF peaks
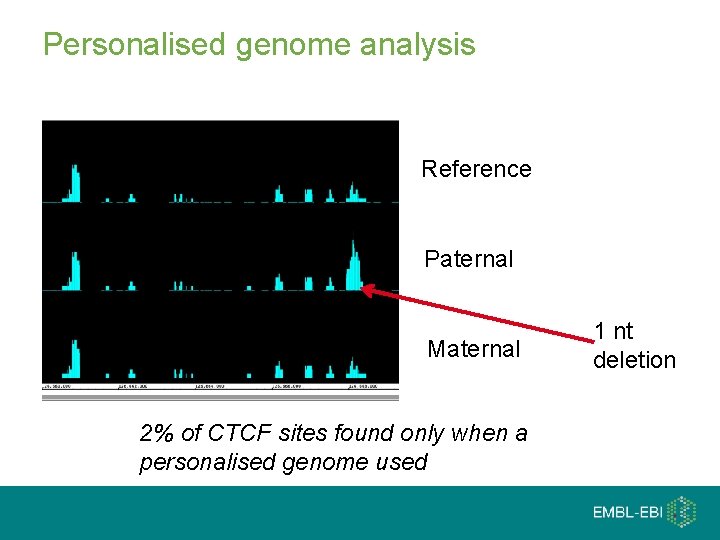
Personalised genome analysis Reference Paternal Maternal 2% of CTCF sites found only when a personalised genome used 1 nt deletion

The ENCODE Consortium Brad Bernstein (Eric Lander, Manolis Kellis, Tony Kouzarides) Ewan Birney (Jim Kent, Mark Gerstein, Bill Noble, Peter Bickel, Ross Hardison, Zhiping Weng) Greg Crawford (Ewan Birney, Jason Lieb, Terry Furey, Vishy Iyer) Jim Kent (David Haussler, Kate Rosenbloom) John Stamatoyannopoulos (Evan Eichler, George Stamatoyannopoulos, Job Dekker, Maynard Olson, Michael Dorschner, Patrick Navas, Phil Green) Mike Snyder (Kevin Struhl, Mark Gerstein, Peggy Farnham, Sherman Weissman) Rick Myers (Barbara Wold) Scott Tenenbaum (Luiz Penalva) Tim Hubbard (Alexandre Reymond, Alfonso Valencia, David Haussler, Ewan Birney, Jim Kent, Manolis Kellis, Mark Gerstein, Michael Brent, Roderic Guigo) Tom Gingeras (Alexandre Reymond, David Spector, Greg Hannon, Michael Brent, Roderic Guigo, Stylianos Antonarakis, Yijun Ruan, Yoshihide Hayashizaki) Zhiping Weng (Nathan Trinklein, Rick Myers) Additional ENCODE Participants: Elliott Marguiles, Eric Green, Job Dekker, Laura Elnitski. . and many senior scientists, postdocs, students, technicians, computer scientists, statisticians and administrators in these groups NHGRI: Elise Feingold, Peter Good 26
Want more? #276 ENCODE whole-genome data at UCSC - New data and access tools #116. PATTERN DISCOVERY AND SEGMENTATION OF CHROMATIN AND RNA DATA IN MULTIPLE CELL LINES #42 Analysis of transcription starting sites with CAGE in the ENCODE project #215 The classification and analysis of lnc. RNAs in GENCODE The Diversity of Human Small RNAs #325 INTEGRATIVE ANALYSIS OF POPULATION DIVERSITY, CONSERVATION, AND EPIGENOMIC INFORMATION AT REGULATORY ELEMENTS AND DISEASEASSOCIATED REGIONS #109 MODULATION OF THE TRANSCRIPTION FACTOR AFFINITY IN THE HUMAN GENOME #285 ABSOLUTE QUANTIFICATION, ERROR MODELLING AND QUALITY CONTROL OF RNASEQ USING SPIKE-IN CONTROL SEQUENCES Plos Biology, April 2011 Another 21 non consortium abstracts use ENCODE data
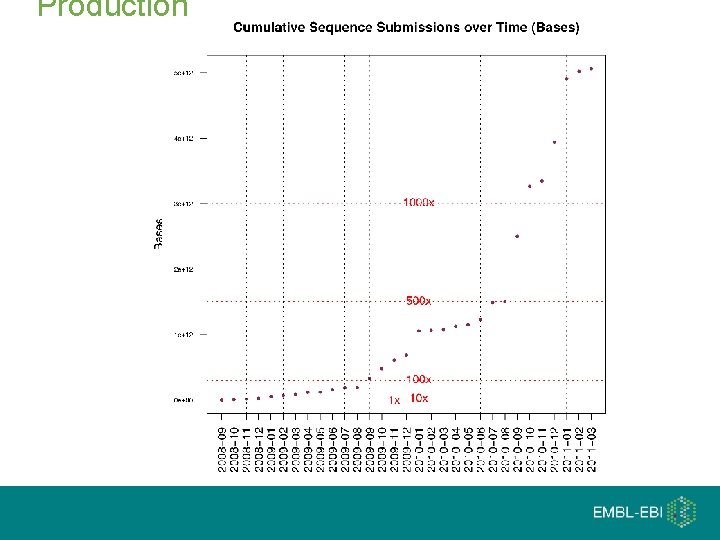
Production
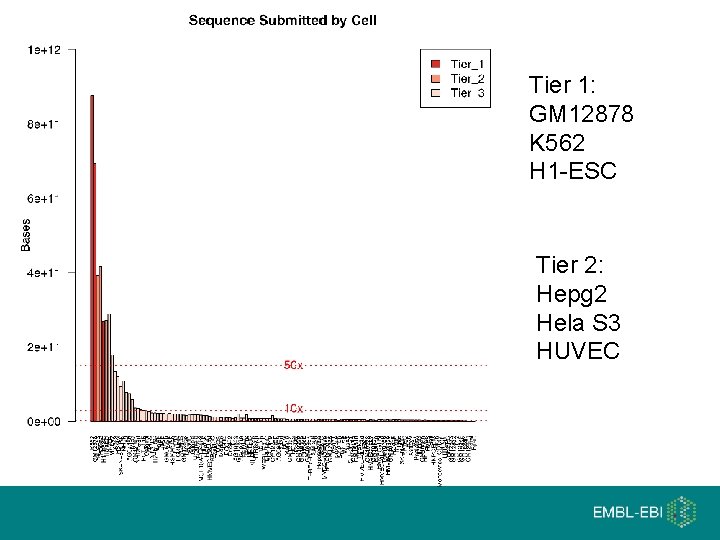
Tier 1: GM 12878 K 562 H 1 -ESC Tier 2: Hepg 2 Hela S 3 HUVEC
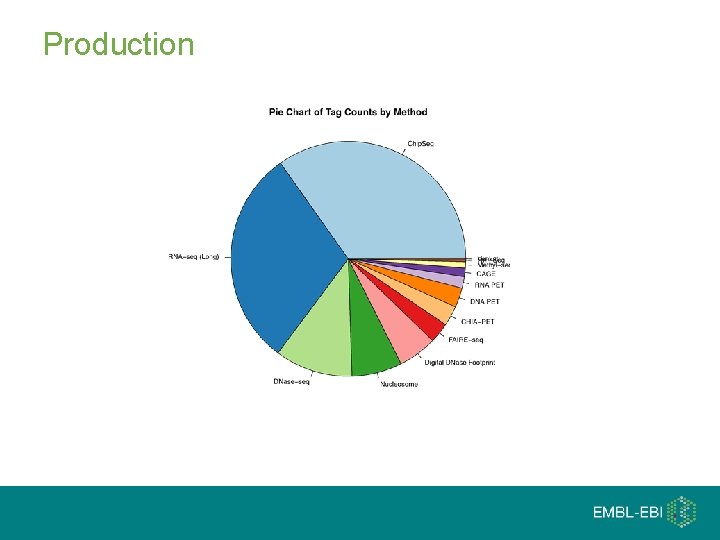
Production
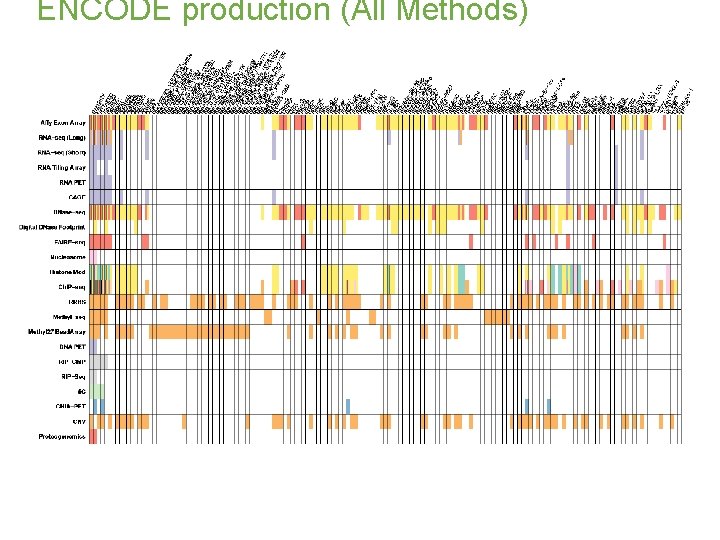
ENCODE production (All Methods)
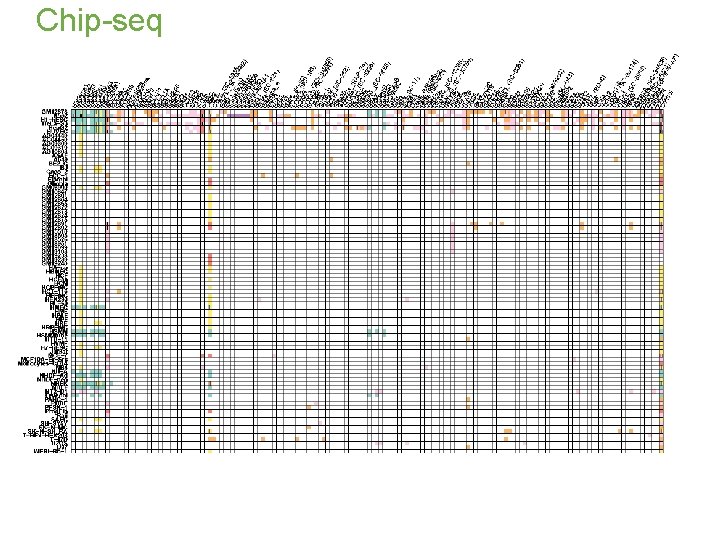
Chip-seq
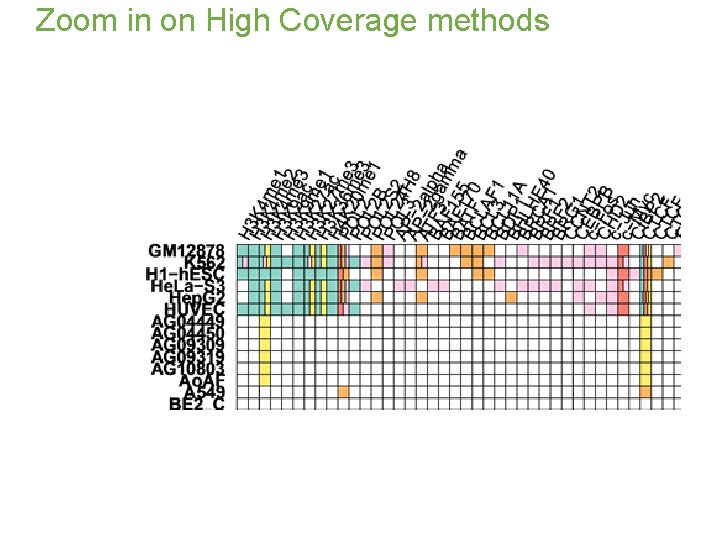
Zoom in on High Coverage methods
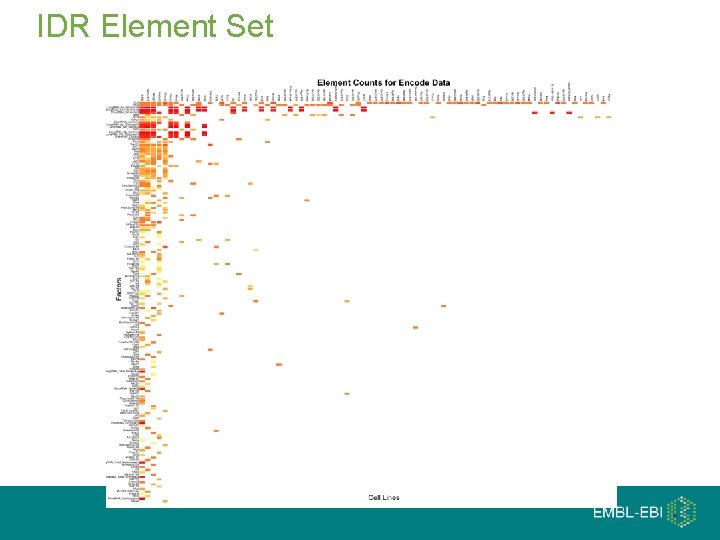
IDR Element Set
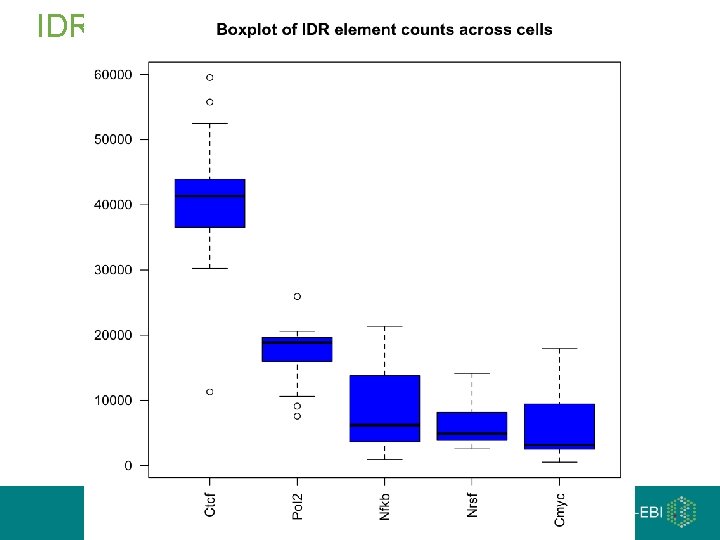
IDR Element Set
Ability to find Enchancers in a directed, discriminative way Sophisticiated (Anshul, chao) Less Sophisticiated (Jim)
- Slides: 36