ENCODE Scientific Presentation July 20 2012 UW ENCODE

- Slides: 27

ENCODE Scientific Presentation July 20, 2012 UW ENCODE group Presenter: John Stamatoyannopoulos

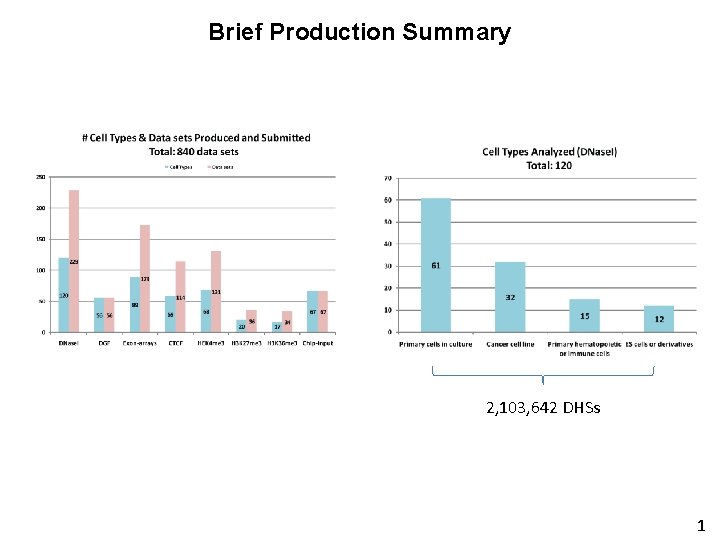
Brief Production Summary 2, 103, 642 DHSs 1

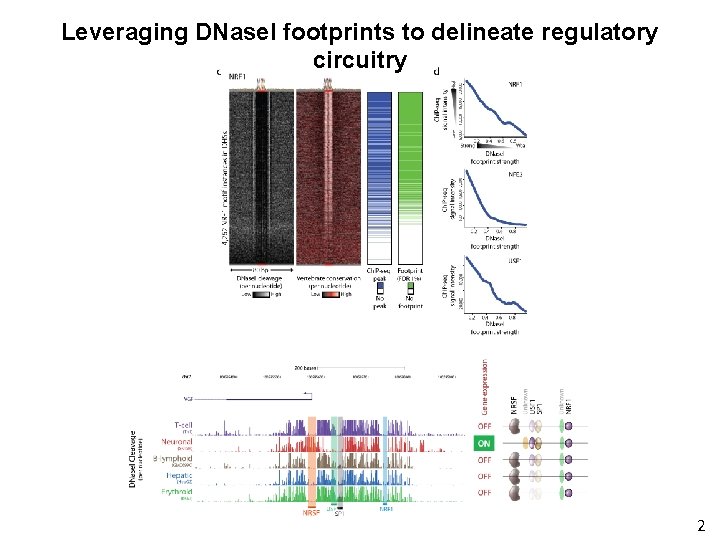
Leveraging DNase. I footprints to delineate regulatory circuitry 2

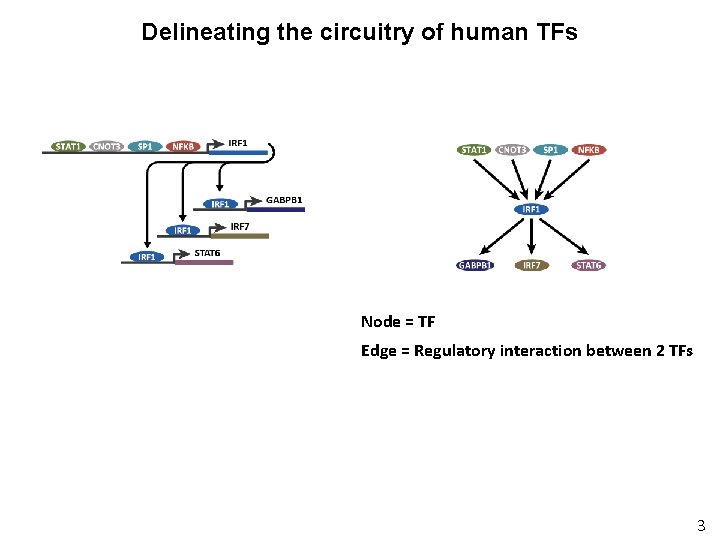
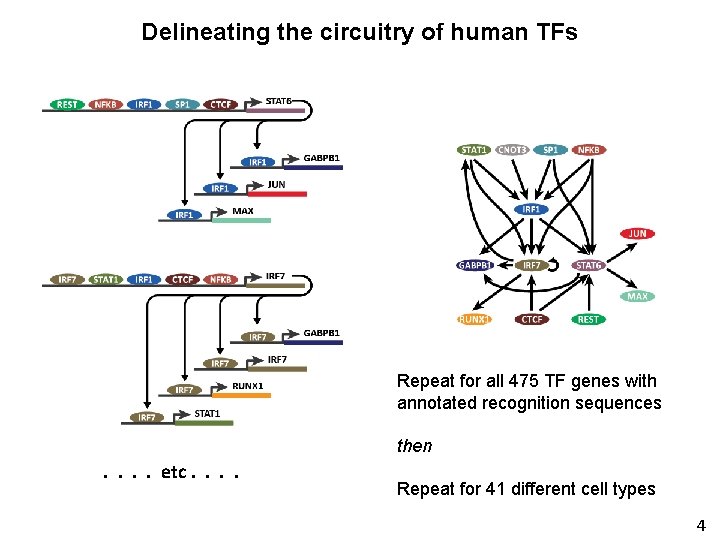
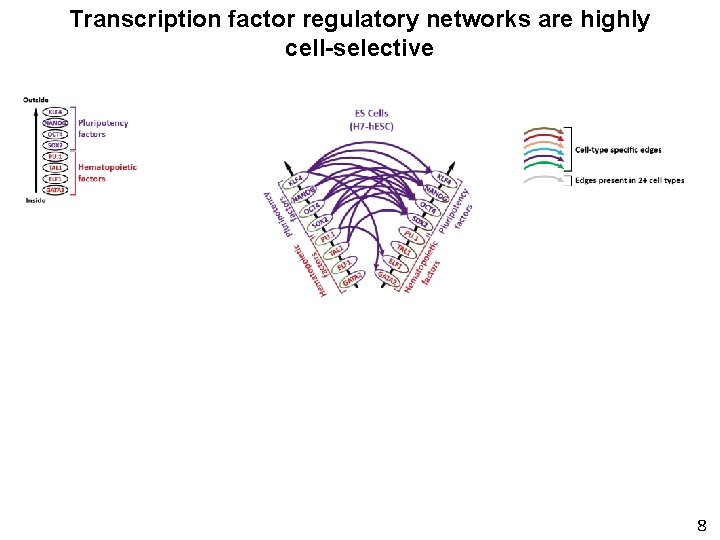
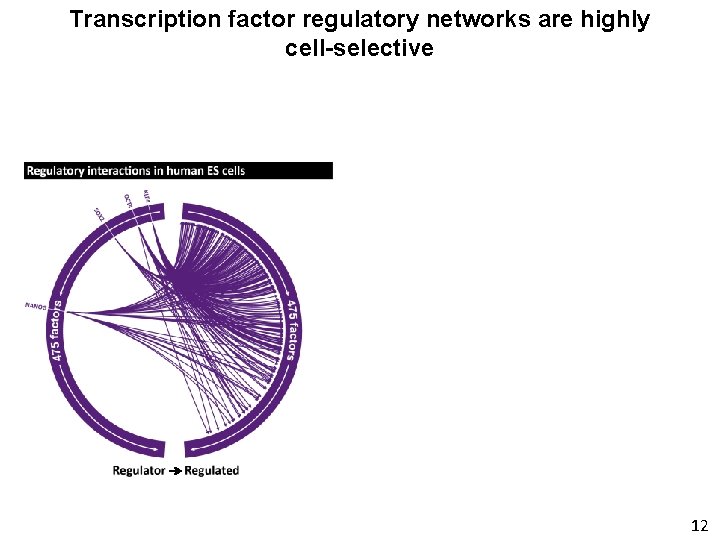
Delineating the circuitry of human TFs Node = TF Edge = Regulatory interaction between 2 TFs 3

Delineating the circuitry of human TFs Repeat for all 475 TF genes with annotated recognition sequences then . . etc. . Repeat for 41 different cell types 4

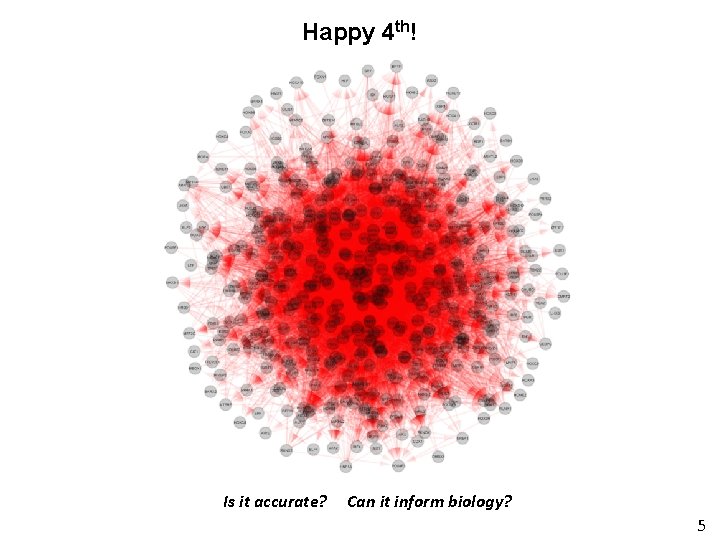
Happy 4 th! Is it accurate? Can it inform biology? 5

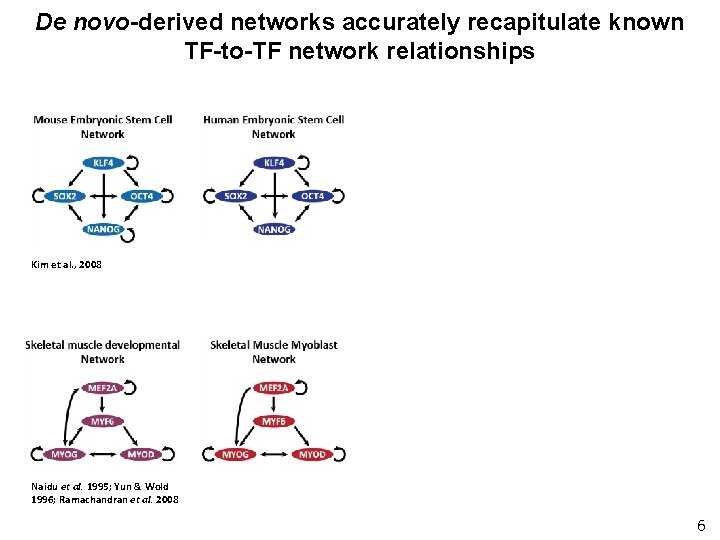
De novo-derived networks accurately recapitulate known TF-to-TF network relationships Kim et al. , 2008 Naidu et al. 1995; Yun & Wold 1996; Ramachandran et al. 2008 6

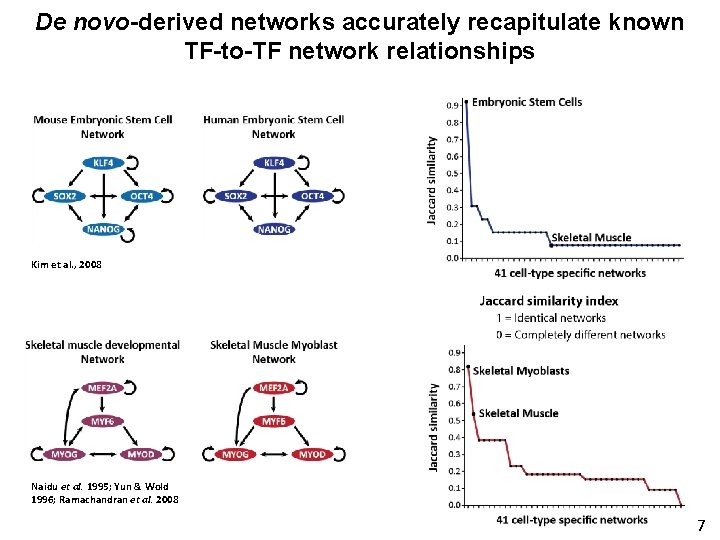
De novo-derived networks accurately recapitulate known TF-to-TF network relationships Kim et al. , 2008 Naidu et al. 1995; Yun & Wold 1996; Ramachandran et al. 2008 7

Transcription factor regulatory networks are highly cell-selective 8

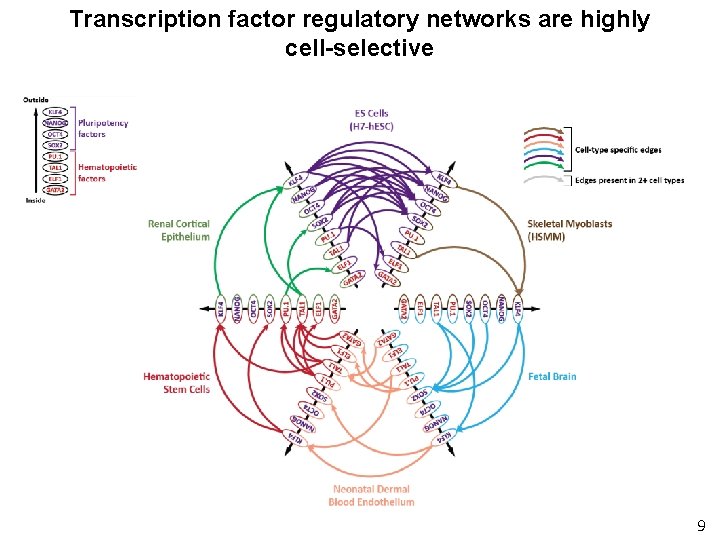
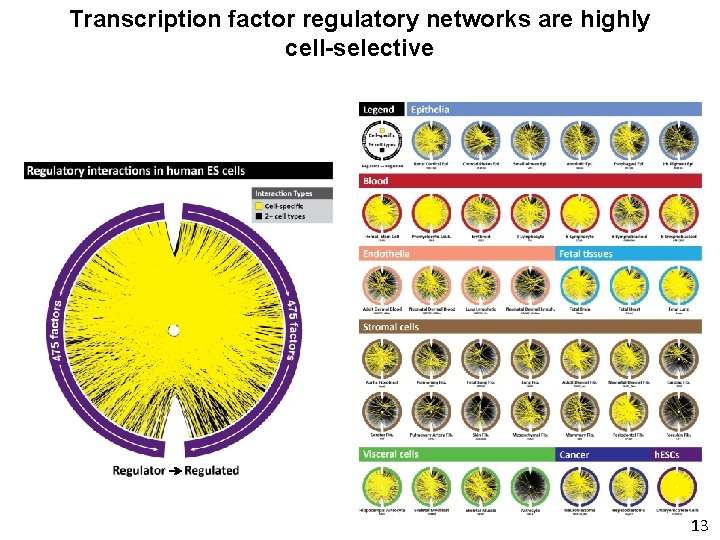
Transcription factor regulatory networks are highly cell-selective 9

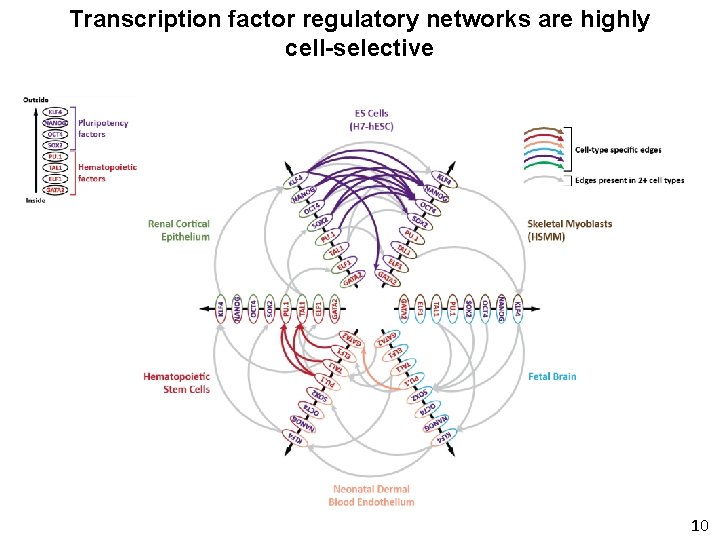
Transcription factor regulatory networks are highly cell-selective 10

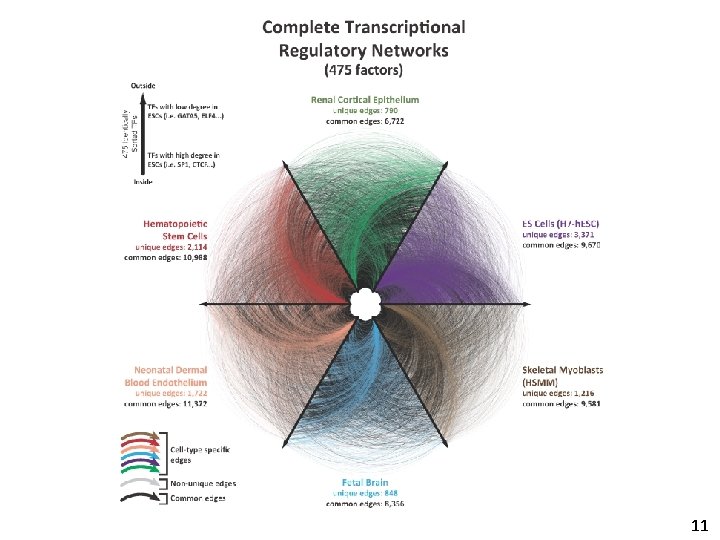
11

Transcription factor regulatory networks are highly cell-selective 12

Transcription factor regulatory networks are highly cell-selective 13

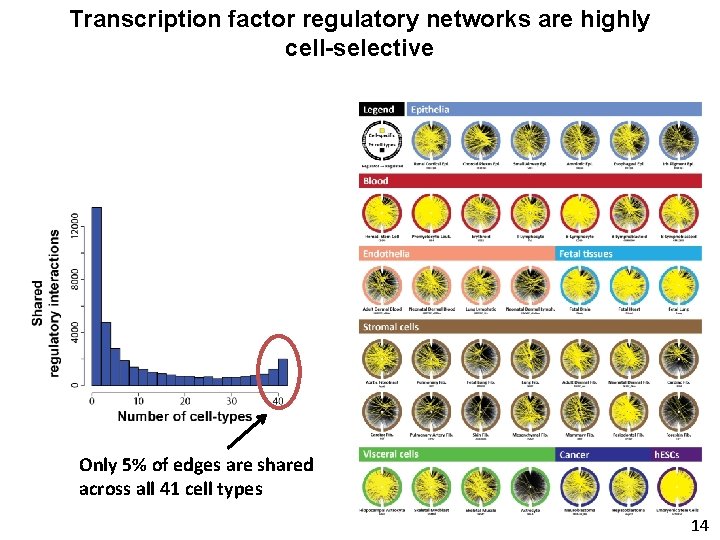
Transcription factor regulatory networks are highly cell-selective Only 5% of edges are shared across all 41 cell types 14

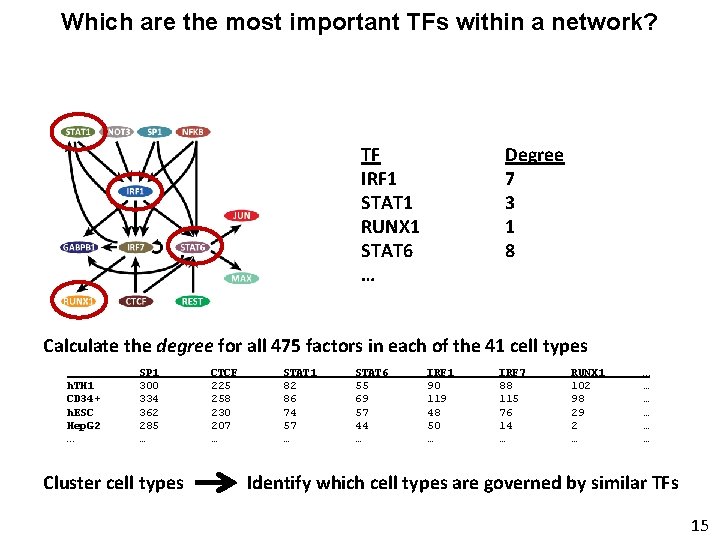
Which are the most important TFs within a network? TF IRF 1 STAT 1 RUNX 1 STAT 6 … Degree 7 3 1 8 Calculate the degree for all 475 factors in each of the 41 cell types h. TH 1 CD 34+ h. ESC Hep. G 2 … SP 1 300 334 362 285 … Cluster cell types CTCF 225 258 230 207 … STAT 1 82 86 74 57 … STAT 6 55 69 57 44 … IRF 1 90 119 48 50 … IRF 7 88 115 76 14 … RUNX 1 102 98 29 2 … … … … Identify which cell types are governed by similar TFs 15

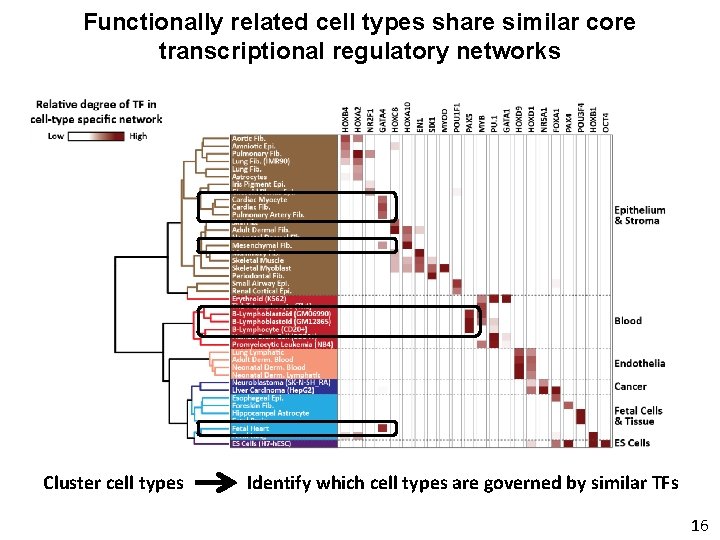
Functionally related cell types share similar core transcriptional regulatory networks Cluster cell types Identify which cell types are governed by similar TFs 16

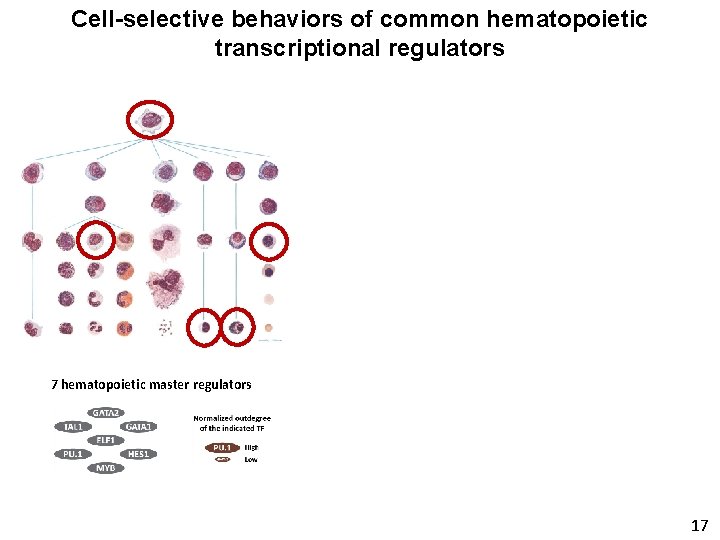
Cell-selective behaviors of common hematopoietic transcriptional regulators 7 hematopoietic master regulators 17

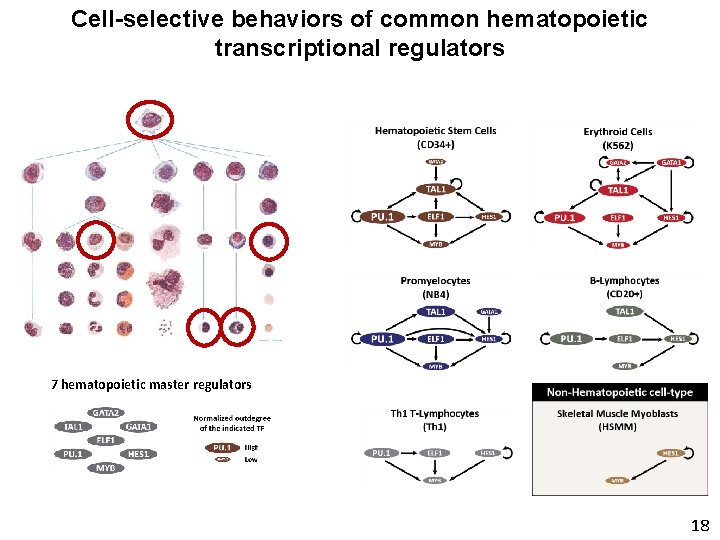
Cell-selective behaviors of common hematopoietic transcriptional regulators 7 hematopoietic master regulators 18

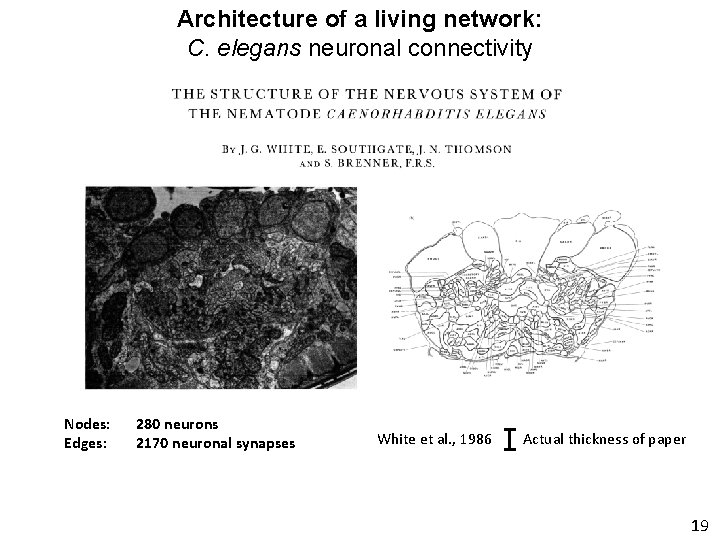
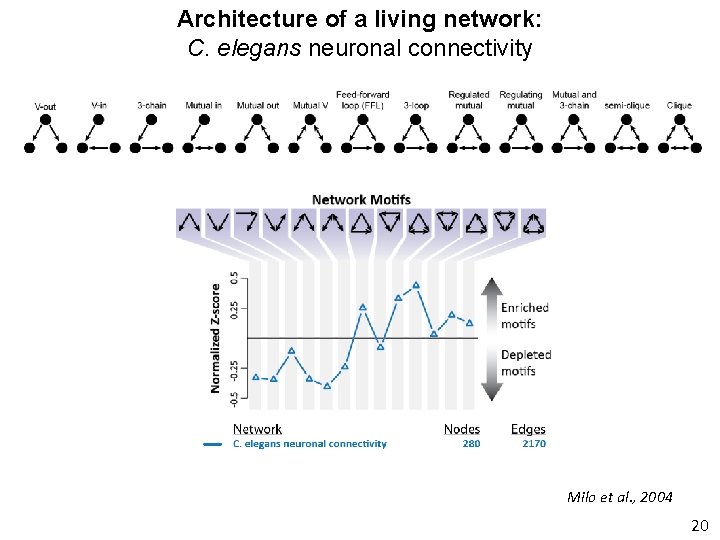
Architecture of a living network: C. elegans neuronal connectivity Nodes: Edges: 280 neurons 2170 neuronal synapses White et al. , 1986 Actual thickness of paper 19

Architecture of a living network: C. elegans neuronal connectivity Milo et al. , 2004 20

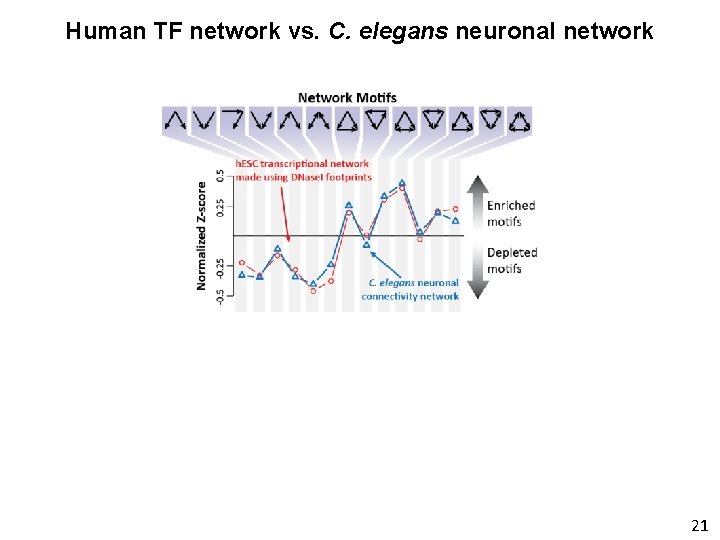
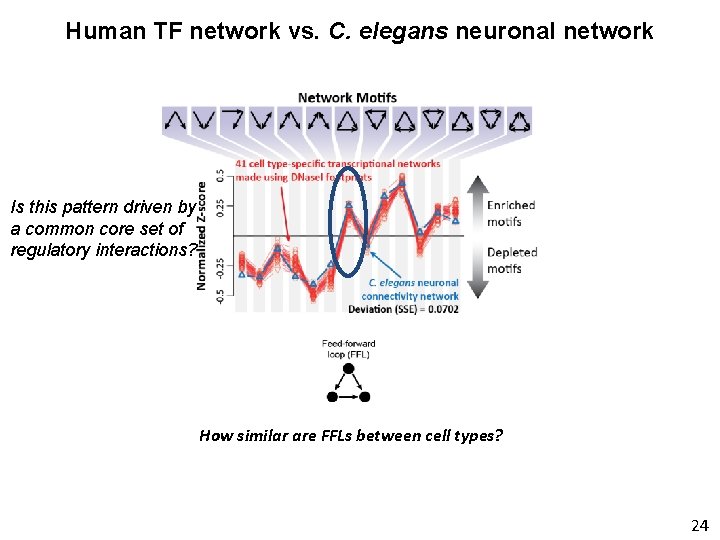
Human TF network vs. C. elegans neuronal network 21

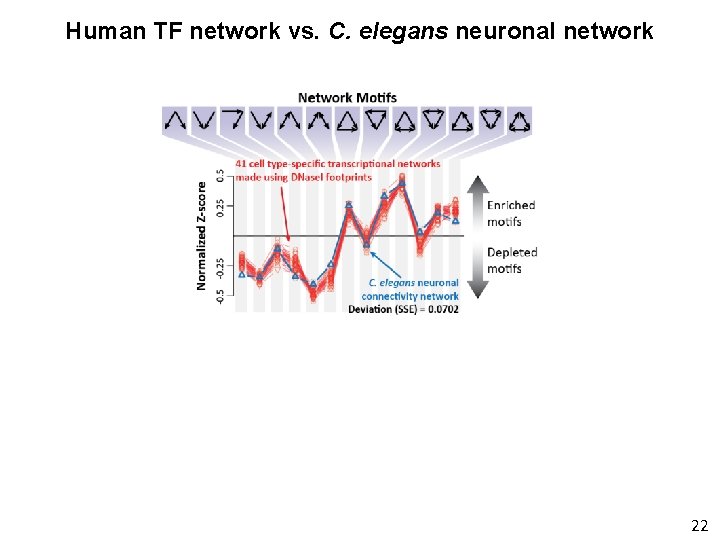
Human TF network vs. C. elegans neuronal network 22

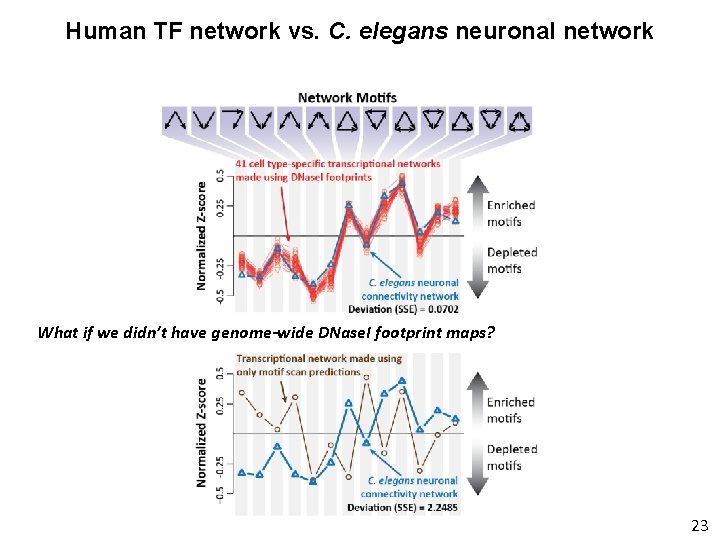
Human TF network vs. C. elegans neuronal network What if we didn’t have genome-wide DNase. I footprint maps? 23

Human TF network vs. C. elegans neuronal network Is this pattern driven by a common core set of regulatory interactions? How similar are FFLs between cell types? 24

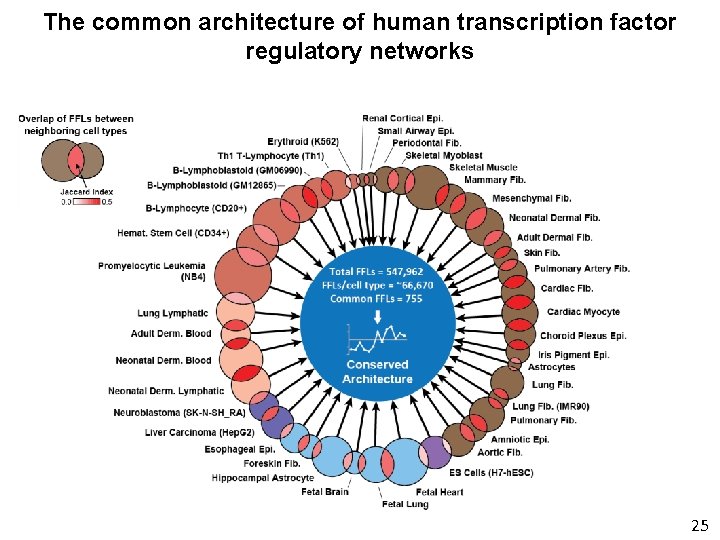
The common architecture of human transcription factor regulatory networks 25

Acknowledgements Network Analysis Shane Neph, Andrew Stergachis, Alex Reynolds, Richard Sandstrom, Elhanan Borenstein Genomic DNase. I footprinting 26