ENCODE Data Available through The UCSC Genome Browser
ENCODE Data Available through The UCSC Genome Browser Osvaldo Graña CNIO Bioinformatics Unit Materials prepared by Mary Mangan, Ph. D. Warren C. Lathe, Ph. D. www. openhelix. com Version 3 1
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org Copyright Open. Helix. No use or reproduction without express written consent 2
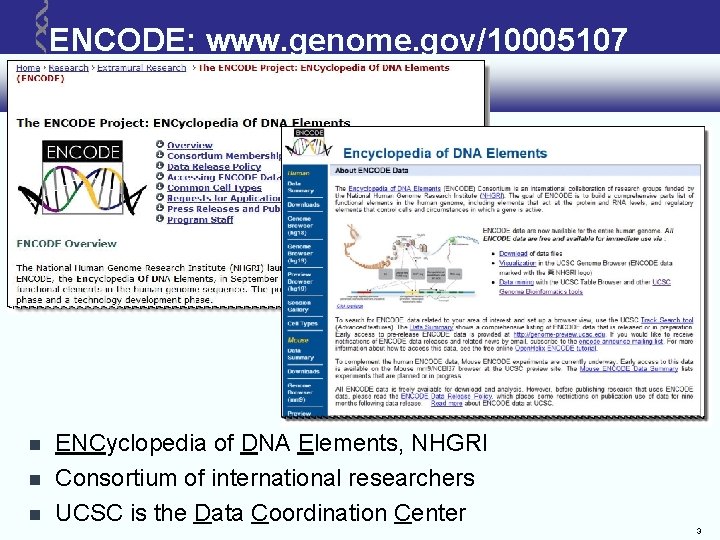
ENCODE: www. genome. gov/10005107 ENCyclopedia of DNA Elements, NHGRI Consortium of international researchers UCSC is the Data Coordination Center 3
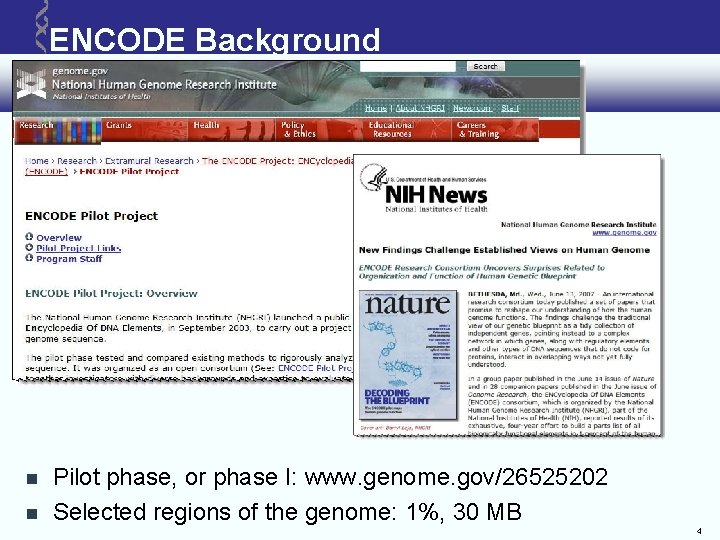
ENCODE Background Pilot phase, or phase I: www. genome. gov/26525202 Selected regions of the genome: 1%, 30 MB 4
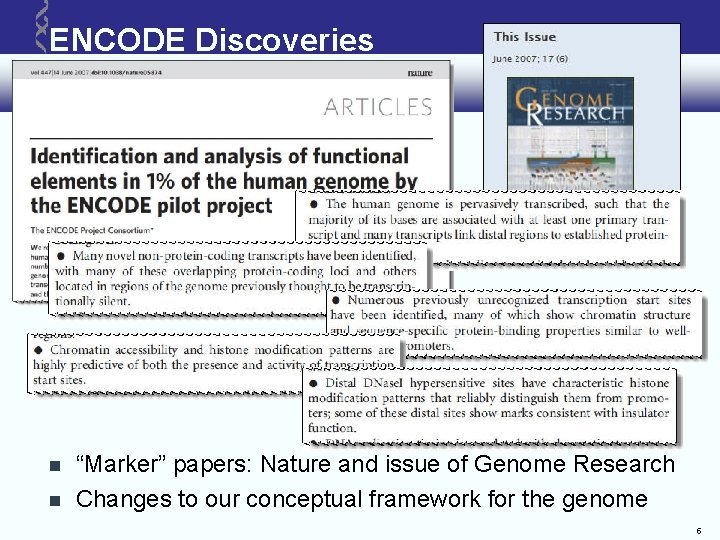
ENCODE Discoveries “Marker” papers: Nature and issue of Genome Research Changes to our conceptual framework for the genome 5
ENCODE Pilot Data and Beyond ENCODE portal: http: //genome. ucsc. edu/ENCODE/ Pilot ENCODE browser: genome. ucsc. edu/ENCODE/pilot. html 6
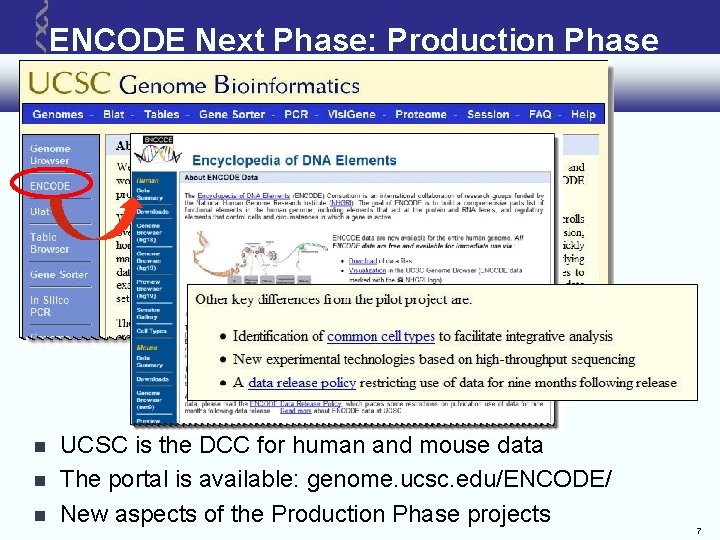
ENCODE Next Phase: Production Phase UCSC is the DCC for human and mouse data The portal is available: genome. ucsc. edu/ENCODE/ New aspects of the Production Phase projects 7
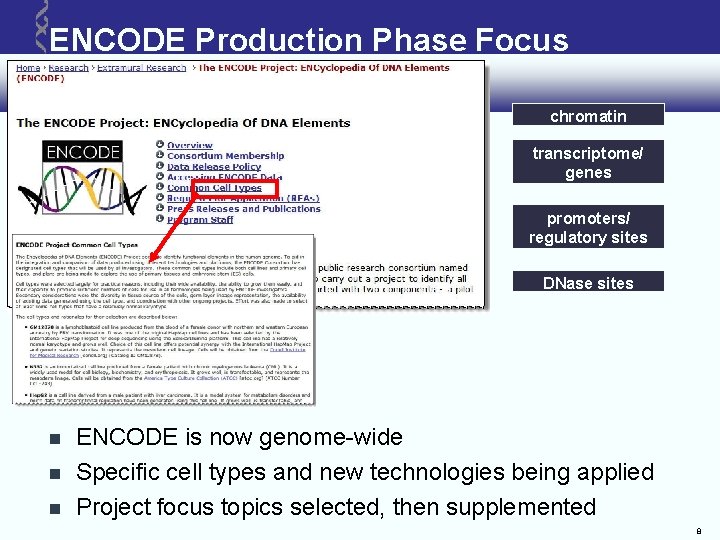
ENCODE Production Phase Focus chromatin transcriptome/ genes promoters/ regulatory sites DNase sites ENCODE is now genome-wide Specific cell types and new technologies being applied Project focus topics selected, then supplemented 8
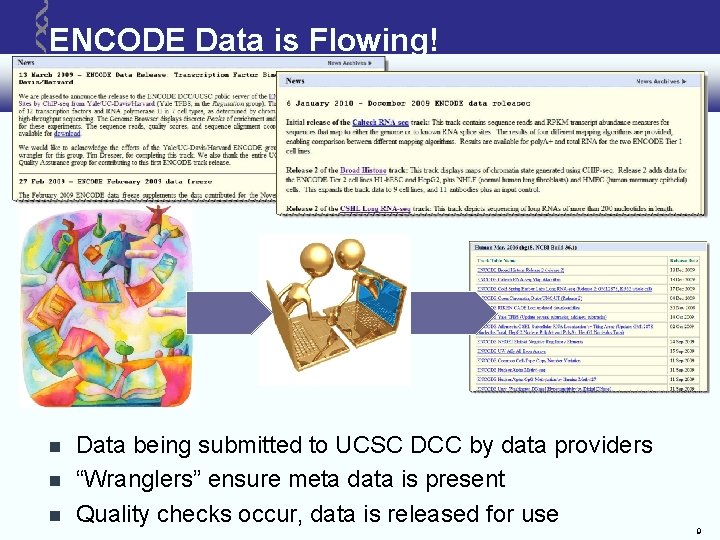
ENCODE Data is Flowing! Data being submitted to UCSC DCC by data providers “Wranglers” ensure meta data is present Quality checks occur, data is released for use 9
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org 10
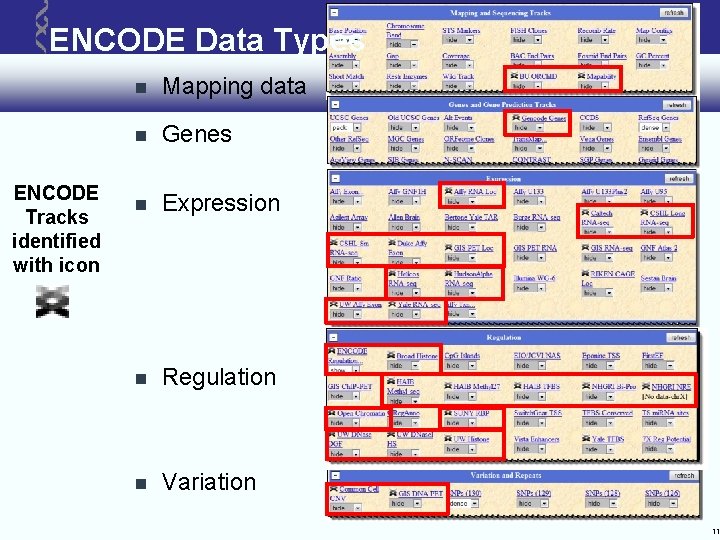
ENCODE Data Types ENCODE Tracks identified with icon Mapping data Genes Expression Regulation Variation 11
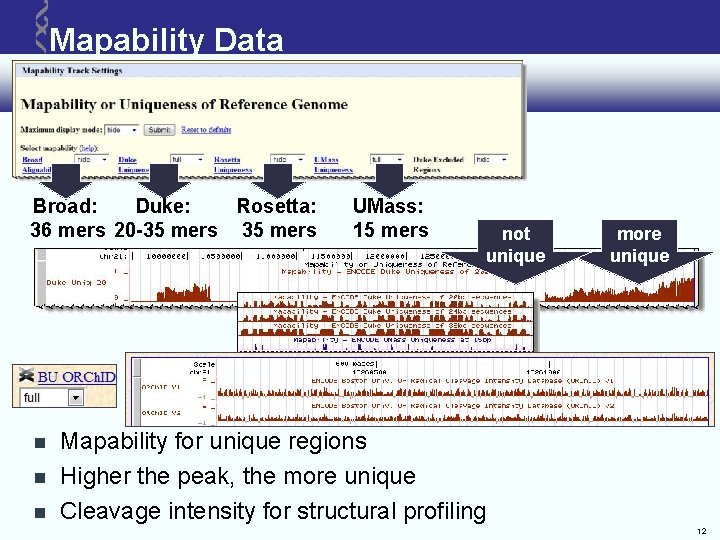
Mapability Data Broad: Duke: Rosetta: 36 mers 20 -35 mers UMass: 15 mers not unique more unique Mapability for unique regions Higher the peak, the more unique Cleavage intensity for structural profiling 12
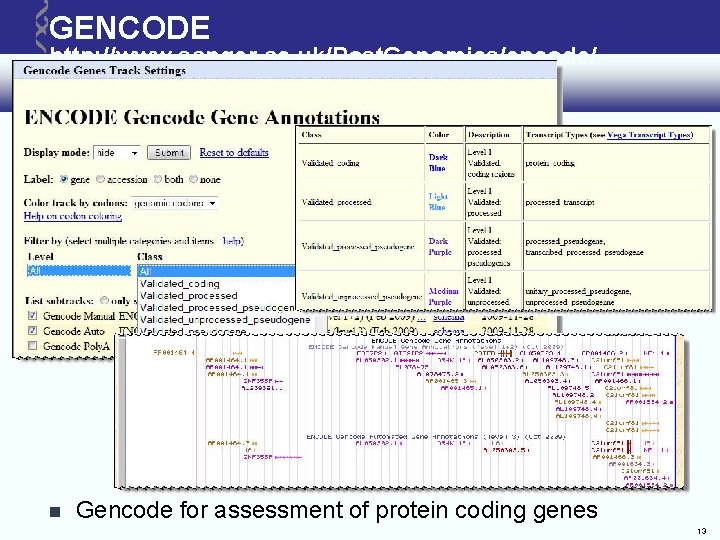
GENCODE http: //www. sanger. ac. uk/Post. Genomics/encode/ Gencode for assessment of protein coding genes 13
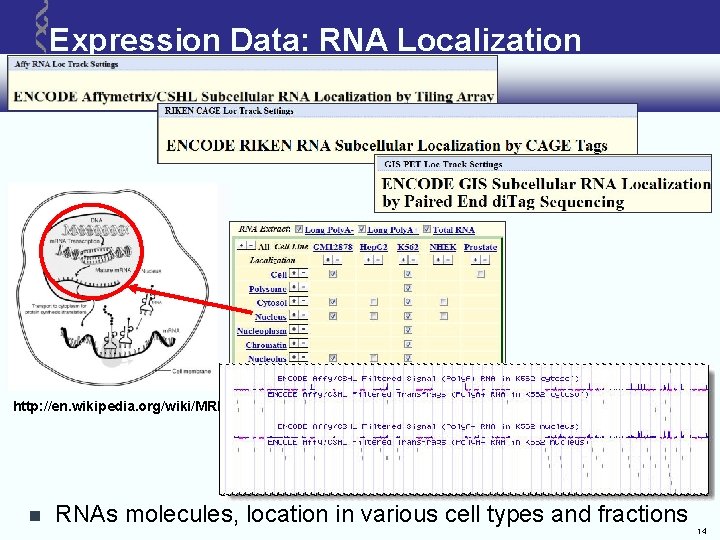
Expression Data: RNA Localization http: //en. wikipedia. org/wiki/MRNA RNAs molecules, location in various cell types and fractions 14
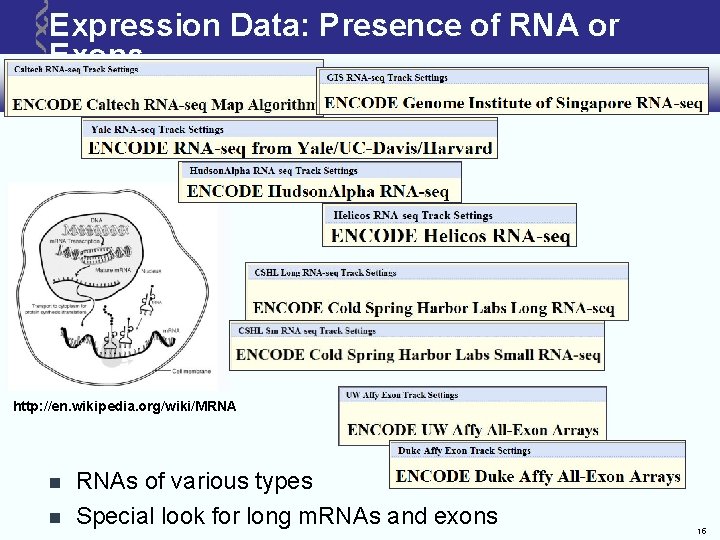
Expression Data: Presence of RNA or Exons http: //en. wikipedia. org/wiki/MRNA RNAs of various types Special look for long m. RNAs and exons 15
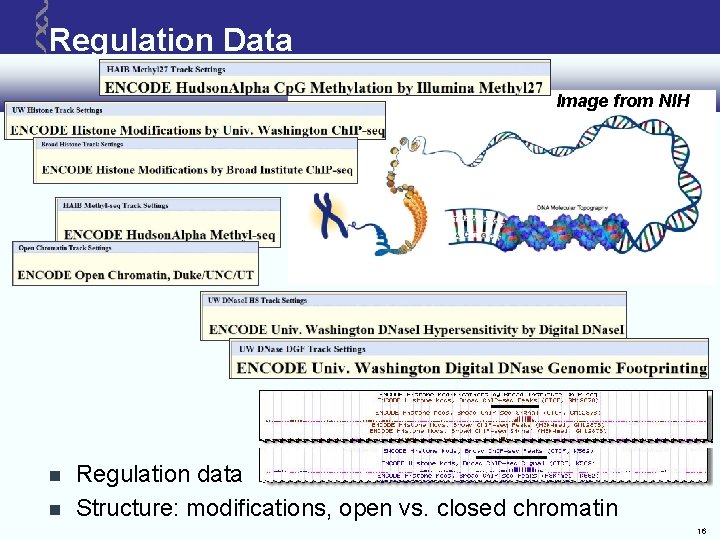
Regulation Data Image from NIH Regulation data Structure: modifications, open vs. closed chromatin 16
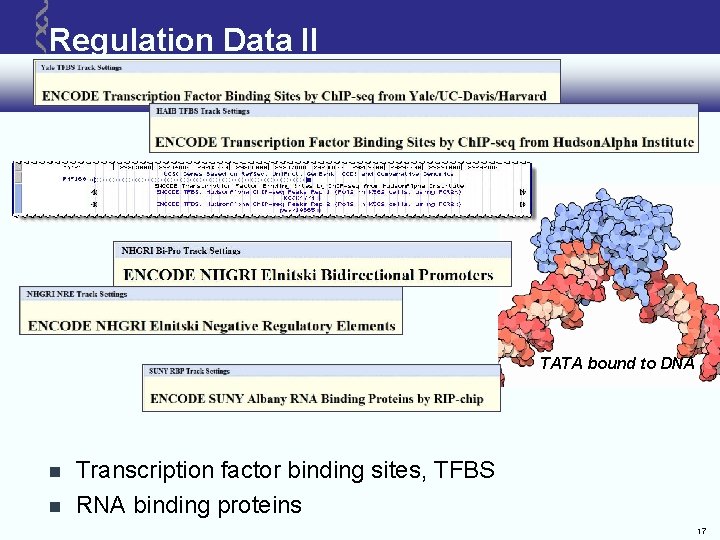
Regulation Data II TATA bound to DNA Transcription factor binding sites, TFBS RNA binding proteins 17
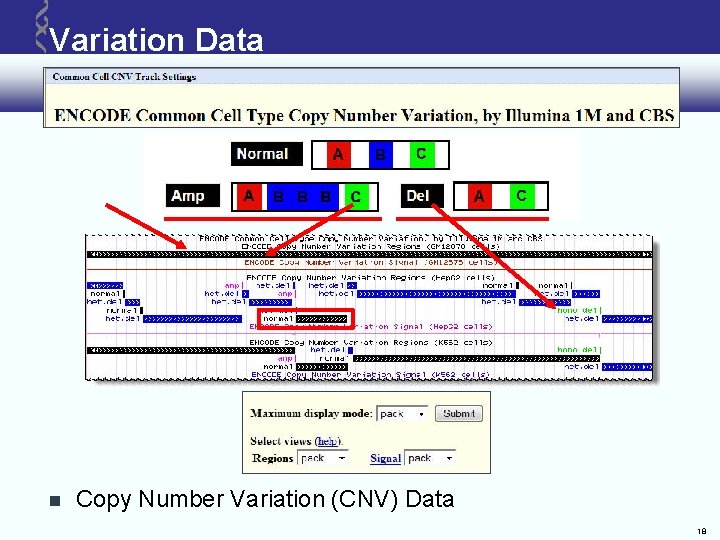
Variation Data Copy Number Variation (CNV) Data 18
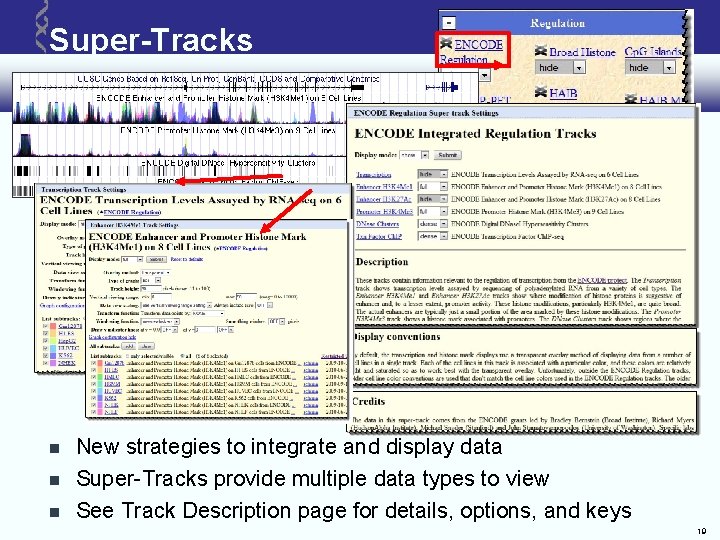
Super-Tracks New strategies to integrate and display data Super-Tracks provide multiple data types to view See Track Description page for details, options, and keys 19
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org 20
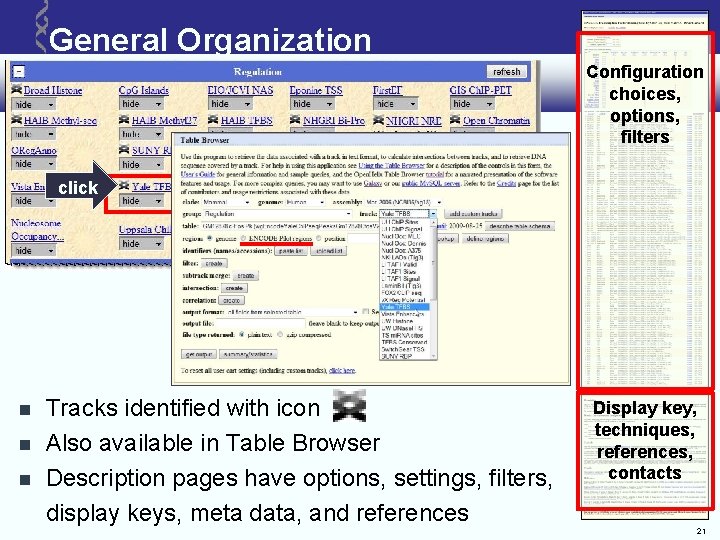
General Organization Configuration choices, options, filters click Tracks identified with icon Also available in Table Browser Description pages have options, settings, filters, display keys, meta data, and references Display key, techniques, references, contacts 21
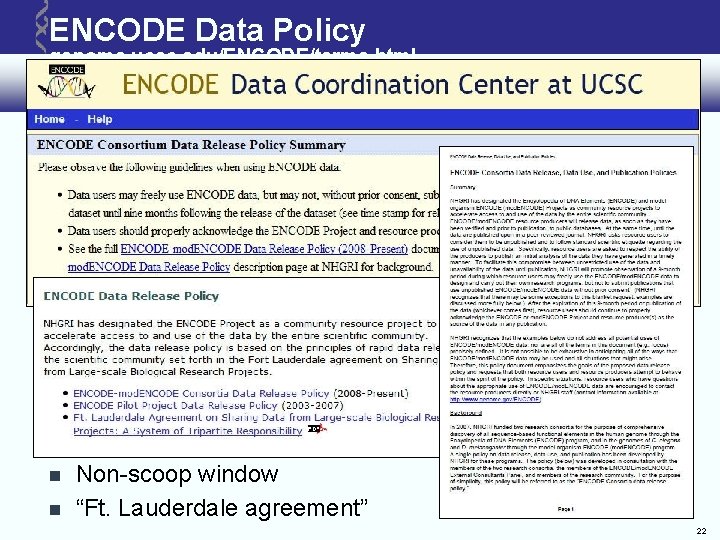
ENCODE Data Policy genome. ucsc. edu/ENCODE/terms. html Non-scoop window “Ft. Lauderdale agreement” 22

Awareness of Embargo Dates Track description pages, Table Browser interface Download pages 23
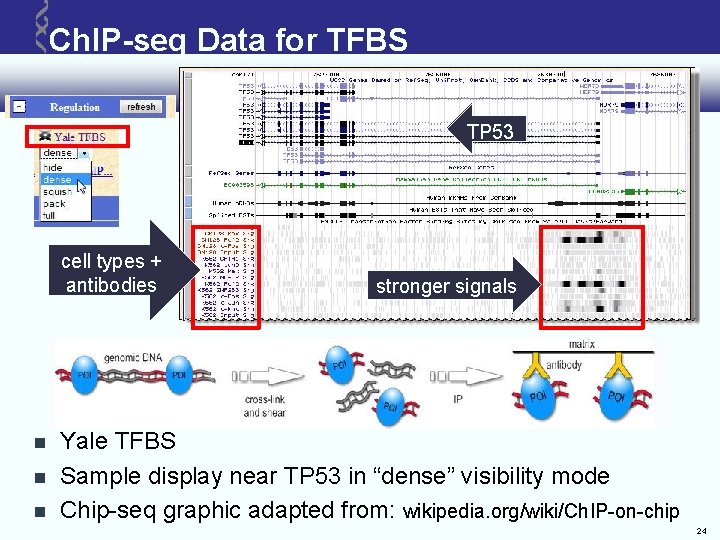
Ch. IP-seq Data for TFBS TP 53 cell types + antibodies stronger signals Yale TFBS Sample display near TP 53 in “dense” visibility mode Chip-seq graphic adapted from: wikipedia. org/wiki/Ch. IP-on-chip 24
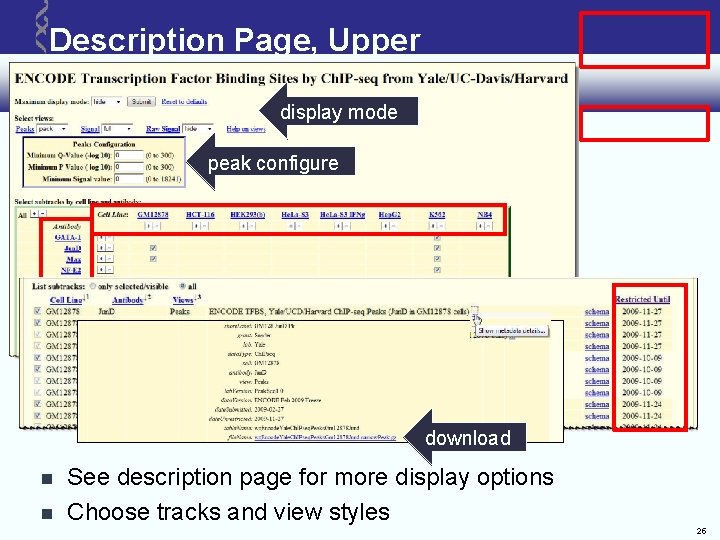
Description Page, Upper display mode peak configure download See description page for more display options Choose tracks and view styles 25
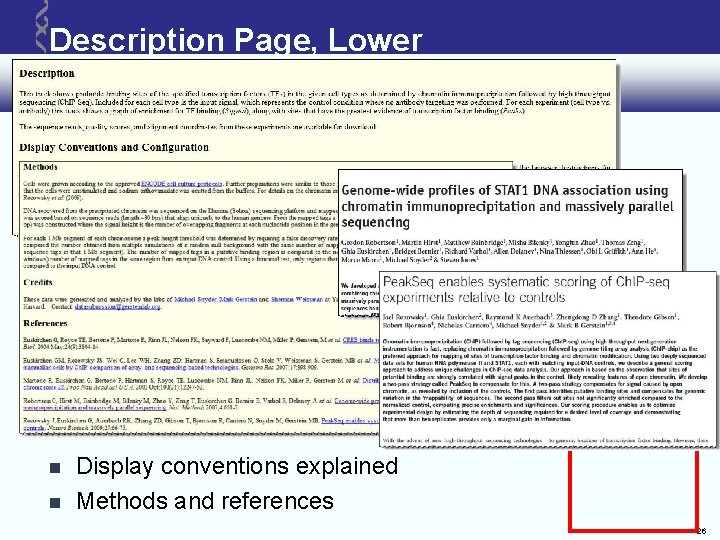
Description Page, Lower Display conventions explained Methods and references 26
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org 27
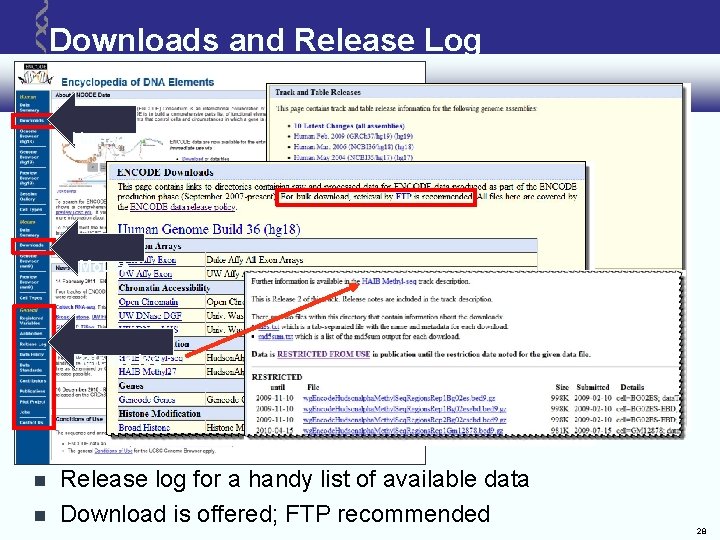
Downloads and Release Log Human Mouse Release log for a handy list of available data Download is offered; FTP recommended 28
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org 29
New Features encode-announce mailing list: https: //lists. soe. ucsc. edu/mailman/listinfo/encode-announce UCSC Genome Browser discussion list: http: //genome. ucsc. edu/contacts. html Mouse data Proteomics data Publications Questions? UCSC mailing list, or ENCODE at NHGRI 30
mod. ENCODE: modencode. org ne w February 2011 issue Science 24 December 2010: Vol. 330 A separate mod. ENCODE: www. genome. gov/26524507 C. elegans and D. melanogaster mod. ENCODE DCC: www. modencode. org 31
ENCODE DCC at UCSC Introduction ENCODE Data Types Find and Use ENCODE Data ENCODE Downloads Additional ENCODE Topics Exercises ENCODE at UCSC: http: //encodeproject. org Copyright Open. Helix. No use or reproduction without express written consent 32

Notice: The materials and slides offered are for non-commercial use only. Reproduction, distribution and/or use for commercial purposes is strictly prohibited. Copyright 2010, Open. Helix, LLC http: //www. openhelix. com/ENCODE Copyright Open. Helix. No use or reproduction without express written consent 33
- Slides: 33