Enabling Clinical Data Reuse with open EHR Data

Enabling Clinical Data Reuse with open. EHR Data Warehouse Environments Luis Marco-Ruiz, Pablo Pazos Gutiérrez, Koray Atalag, Johan Gustav Bellika, Kassaye Yitbarek Yigzaw

Agenda 1. Background 1. 1. Learning Healthcare System 1. 2. Semantic Interoperability 1. 3. Linkage EHR – Inference models 2. METL 2. 1. Modelling 2. 2. Extract 2. 3. Transform 2. 4. Load 3. Experiences 3. 1. Laboratory Service at University Hospital North Norway 3. 2. NZ Cardiac Registry 3. 3. Path based queries 2
Background – Limitations of EBM • Since its inception EBM has improved healthcare outcomes by “collating studies, setting methodologies and publication standards, developing reasons and courses for technical appraisal and building new knowledge bases to be implemented in routine care”[1] • However, some factors like the over-influence of industry in clinical research, the overwhelming amount of evidence in a form of scientific papers, the reduction of knowledge to algorithmic rules and the poor adoption to the individual patient needs have raised as EBM limitations [1] T. Greenhalgh, J. Howick, N. Maskrey, and for the Evidence Based Medicine Renaissance Group, “Evidence based medicine: a movement in crisis? , ” BMJ, vol. 348, no. jun 13 4, pp. g 3725–g 3725, 2014. 3

Background – The Learning Healthcare System • In response to the limitations, the US Institute of medicine (IOM) summarized the pillars needed to overcome them in the proposal of a new healthcare paradigm named the Learning Healthcare System [1]: – (a) fast progression of knowledge produced in clinical research to its use in routine clinical practice; – (b) empowerment of a shared responsibility culture; – (c) present the notion of clinical data as a public asset; – (d) empower interoperability with Patient Health Records (PHR) systems; – (e) facilitate public engagement of patients and doctors. 4

Background – The Learning Healthcare System • The LHS needs efficient data reuse mechanisms that allow to test hypothesis and confirm effects of interventions. • Data need to flow from systems where originally were captured (EHRs, journals, LIS etc. ) to systems that implement inference models (CDS, data analysis etc. ) • Need to find better mechanisms to improve accessibility and processing of clinical data for reuse 5

Background – Ingredients for data reuse • Semantic Interoperability (Si. Op) – Latest efforts (Europe, US, Brazil, etc. ) have established mechanisms to support the adoption of health interoperability standards – Several standards available: open. EHR, HL 7 CDA, ISO 13606, FHIR • Linkage of EHR with inference models – The ‘impedance mismatch’ between the information and inference model needs to be resolved – Mechanisms are needed to rise the level of abstraction of the fine grained data in the EHR to the abstract concepts referenced from inference models (medical logic or data analysis) – Examples: DW, KDOM, Archetype layers, VMR etc. • Data reuse pipeline infrastructure – An infrastructure must adequately implement the mechanisms to resolve the impedance mismatch between EHR and inference models. It must ensure that data is appropriately updated, validated and accessible at the end of the pipeline for reuse. 6
Semantic Interoperability • Integration and harmonization of formats using health information standards – open. EHR, HL 7 CDA, CIMI, FHIR etc. • Definition of shared information models and terminology binding – Several national and international initiatives: Norwegian open. EHR CKM, Spanish ISO 13606 SOM, International open. EHR CKM, CIMI etc. 7
Linkage of EHR with inference models • The ‘impedance mismatch’ between the information and inference model needs to be resolved • Mechanisms to rise the level of abstraction of the fine grained data in the EHR to the abstract concepts referenced from inference models (medical logic or data analysis) • Examples: DW, KDOM, Archetype layers, VMR etc. 8
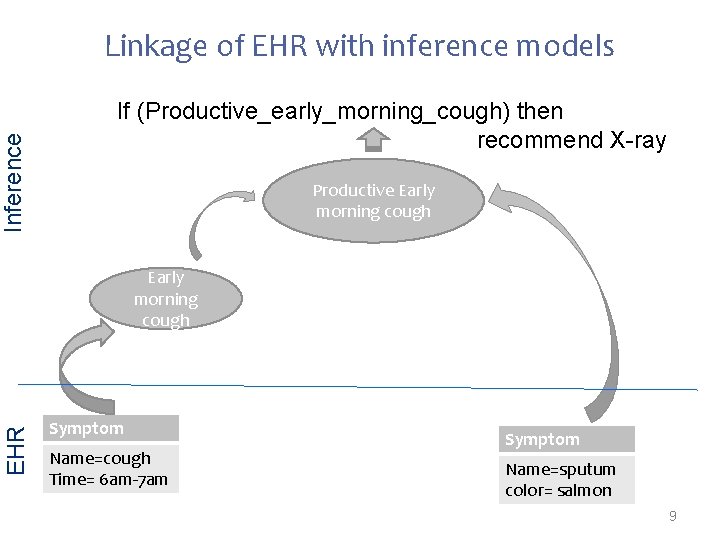
Inference Linkage of EHR with inference models If (Productive_early_morning_cough) then recommend X-ray Productive Early morning cough EHR Early morning cough Symptom Name=cough Time= 6 am-7 am Symptom Name=sputum color= salmon 9
Data reuse pipeline infrastructure • An infrastructure must adequately implement the mechanisms to resolve the ‘impedance mismatch’ between EHR and inference models • It must ensure that data is appropriately updated, validated and accessible at the end of the pipeline for reuse • Transformation from proprietary to EHR standards is the most complex step • The data model must be generic to ensure that the maximum reuse scenarios are covered 10
Challenge To define a infrastructure that appropriately enables: – To gather proprietary clinical data and transform it into standard compliant canonical form (ensures Si. Op) – To query data referencing standard defined clinical models independently from the underlying technological implementation – To define different views of the open. EHR data for different scenarios 11

Agenda 1. Background 1. 1. Learning Healthcare System 1. 2. Semantic Interoperability 1. 3. Linkage EHR – Inference models 2. METL 2. 1. Modelling 2. 2. Extract 2. 3. Transform 2. 4. Load 3. Experiences 3. 1. Laboratory Service at University Hospital North Norway 3. 2. NZ Cardiac Registry 3. 3. Path based queries 12

METL
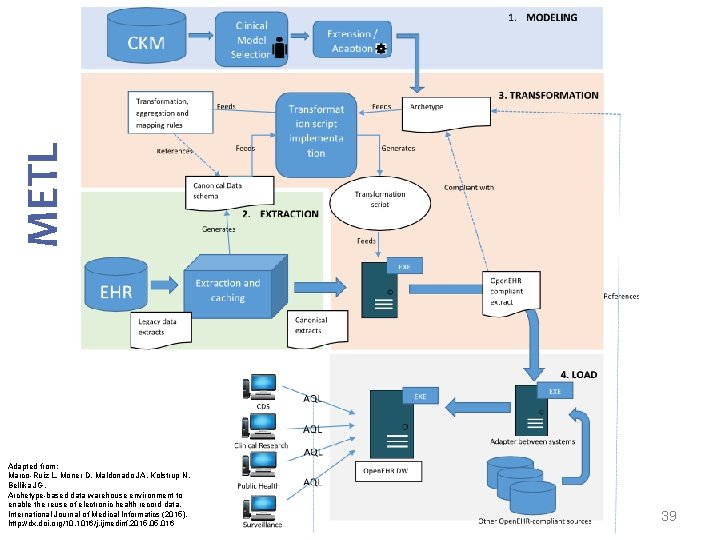
METL Modelling Extract Transform Load Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 14
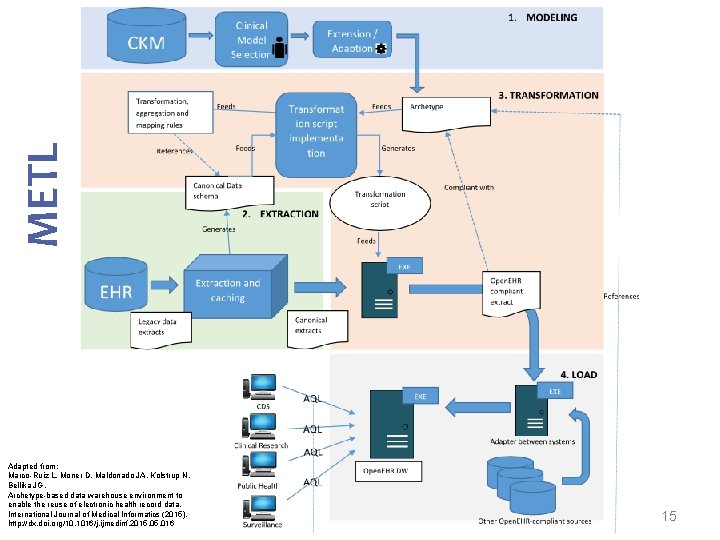
METL Adapted from: Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 15
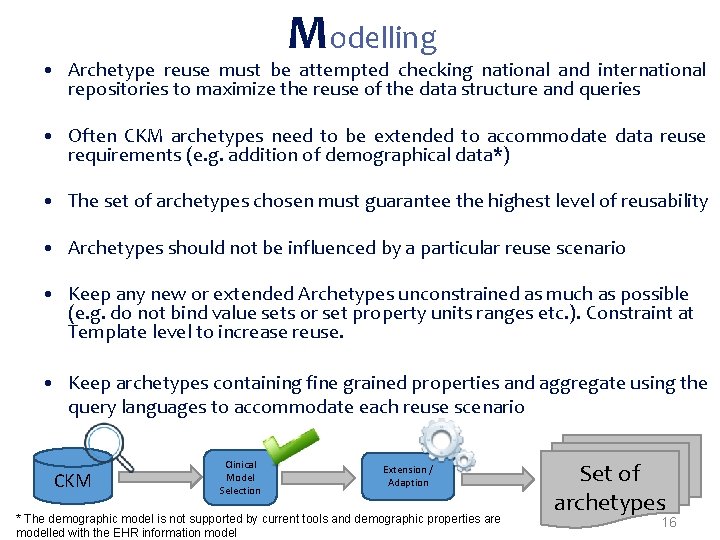
Modelling • Archetype reuse must be attempted checking national and international repositories to maximize the reuse of the data structure and queries • Often CKM archetypes need to be extended to accommodate data reuse requirements (e. g. addition of demographical data*) • The set of archetypes chosen must guarantee the highest level of reusability • Archetypes should not be influenced by a particular reuse scenario • Keep any new or extended Archetypes unconstrained as much as possible (e. g. do not bind value sets or set property units ranges etc. ). Constraint at Template level to increase reuse. • Keep archetypes containing fine grained properties and aggregate using the query languages to accommodate each reuse scenario CKM Clinical Model Selection Extension / Adaption * The demographic model is not supported by current tools and demographic properties are modelled with the EHR information model Set of archetypes 16

Extract High level architectures for Extraction to DW Extraction of data in the Snow system 17
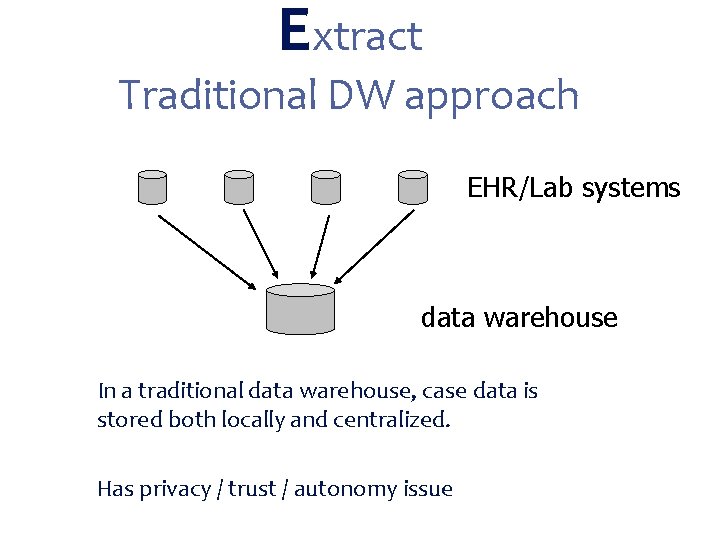
Extract Traditional DW approach EHR/Lab systems data warehouse In a traditional data warehouse, case data is stored both locally and centralized. Has privacy / trust / autonomy issue
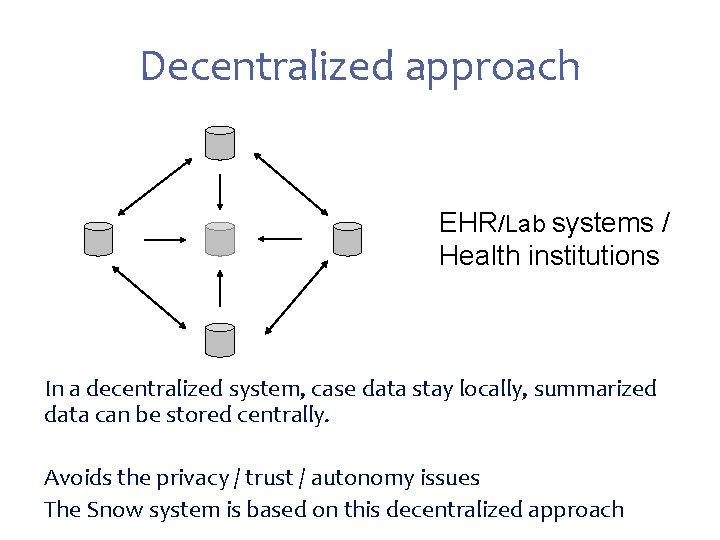
Decentralized approach EHR/Lab systems / Health institutions In a decentralized system, case data stay locally, summarized data can be stored centrally. Avoids the privacy / trust / autonomy issues The Snow system is based on this decentralized approach
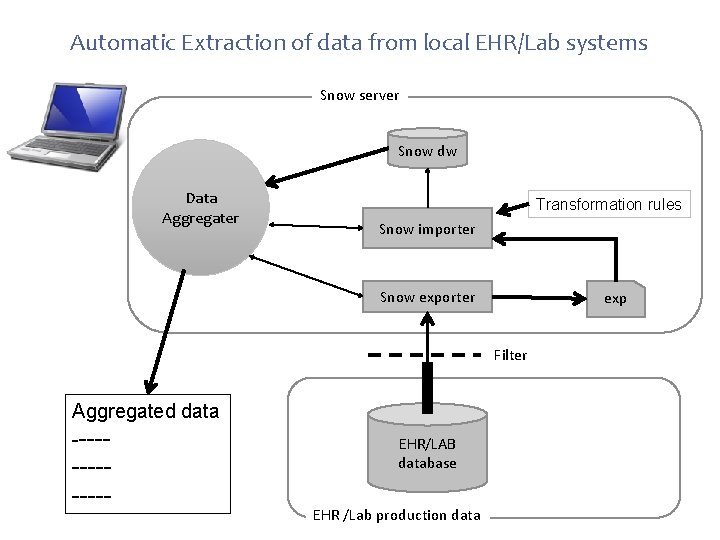
Automatic Extraction of data from local EHR/Lab systems Snow server Snow dw Data Aggregater Transformation rules Snow importer Snow exporter exp Filter Aggregated data --------- EHR/LAB database EHR /Lab production data
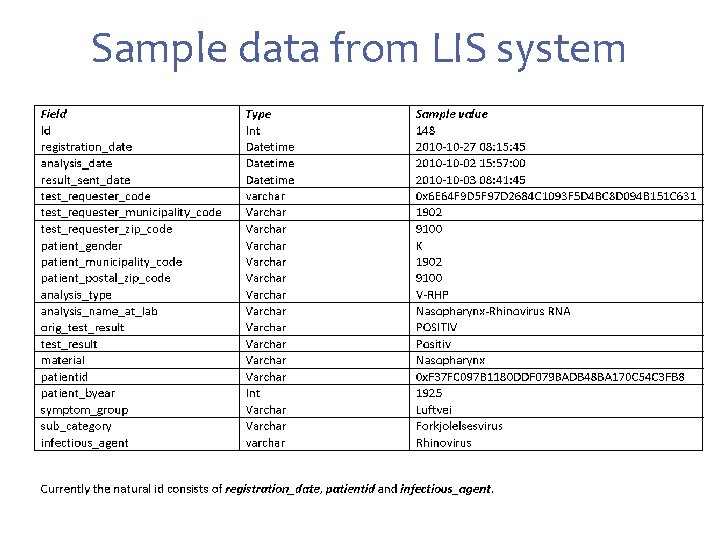
Sample data from LIS system
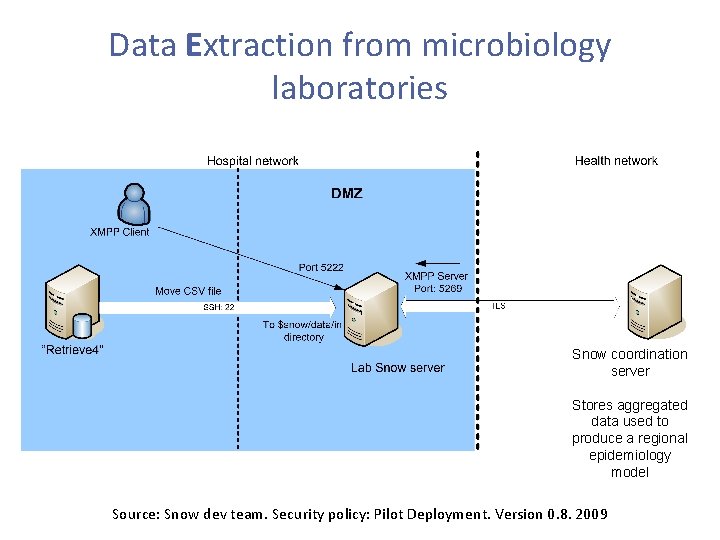
Data Extraction from microbiology laboratories Snow coordination server Stores aggregated data used to produce a regional epidemiology model Source: Snow dev team. Security policy: Pilot Deployment. Version 0. 8. 2009
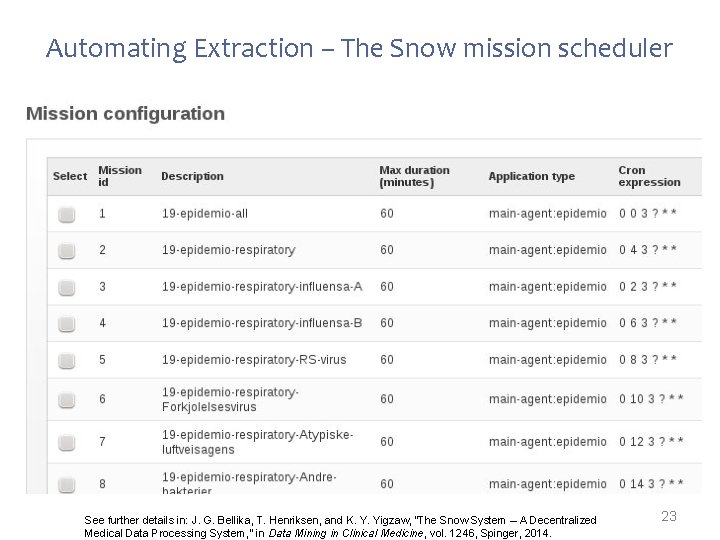
Automating Extraction – The Snow mission scheduler See further details in: J. G. Bellika, T. Henriksen, and K. Y. Yigzaw, “The Snow System – A Decentralized Medical Data Processing System, ” in Data Mining in Clinical Medicine, vol. 1246, Spinger, 2014. 23
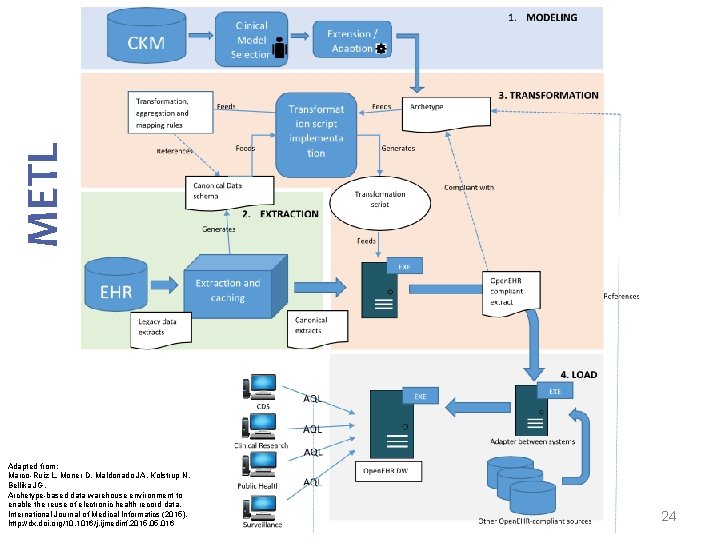
METL Adapted from: Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 24
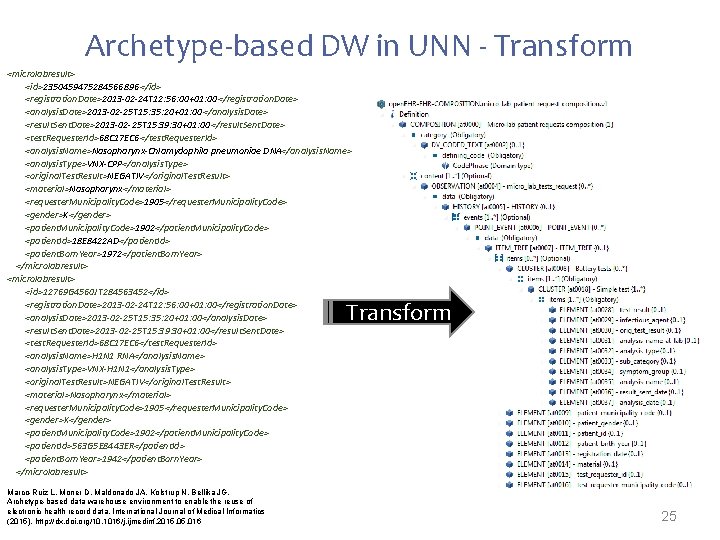
Archetype-based DW in UNN - Transform <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Nasopharynx-Chlamydophila pneumoniae DNA</analysis. Name> <analysis. Type>VNX-CPP</analysis. Type> <original. Test. Result>NEGATIV</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>18 E 8422 AD</patient. Id> <patient. Born. Year>1972</patient. Born. Year> </microlabresult> <id>12769 G 4560 JT 284563452</id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>H 1 N 1 RNA</analysis. Name> <analysis. Type>VNX-H 1 N 1</analysis. Type> <original. Test. Result>NEGATIV</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>563 G 5 E 8443 ER</patient. Id> <patient. Born. Year>1942</patient. Born. Year> </microlabresult> Transform Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 25
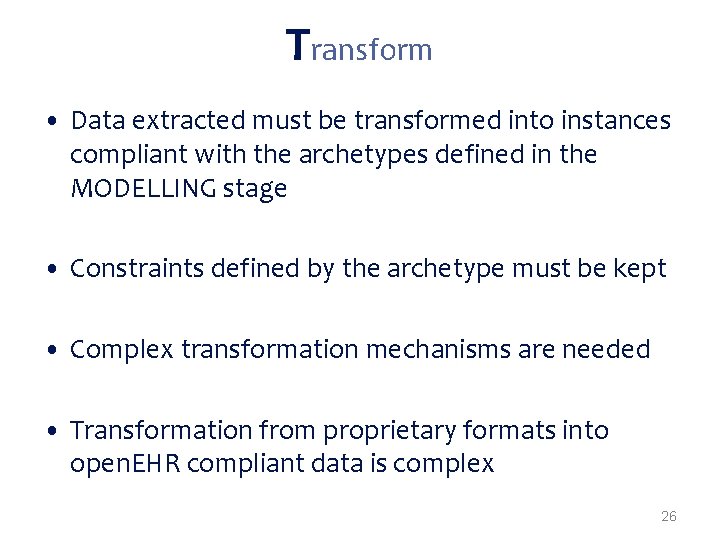
Transform • Data extracted must be transformed into instances compliant with the archetypes defined in the MODELLING stage • Constraints defined by the archetype must be kept • Complex transformation mechanisms are needed • Transformation from proprietary formats into open. EHR compliant data is complex 26
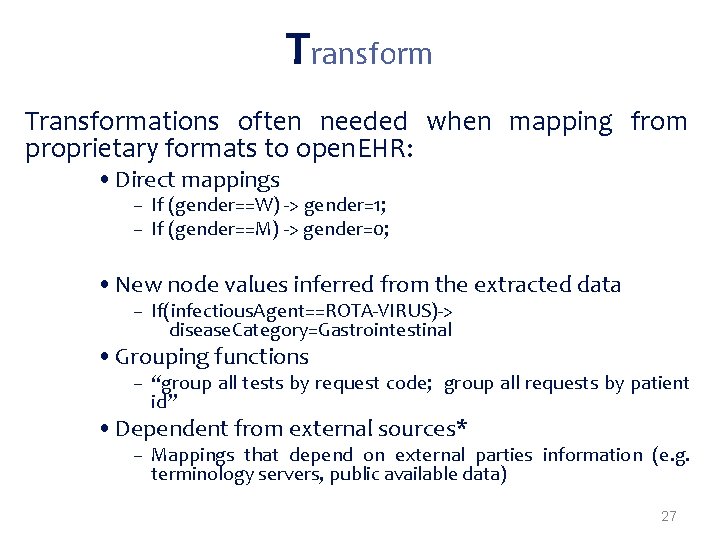
Transformations often needed when mapping from proprietary formats to open. EHR: • Direct mappings – If (gender==W) -> gender=1; – If (gender==M) -> gender=0; • New node values inferred from the extracted data – If(infectious. Agent==ROTA-VIRUS)-> disease. Category=Gastrointestinal • Grouping functions – “group all tests by request code; group all requests by patient id” • Dependent from external sources* – Mappings that depend on external parties information (e. g. terminology servers, public available data) 27
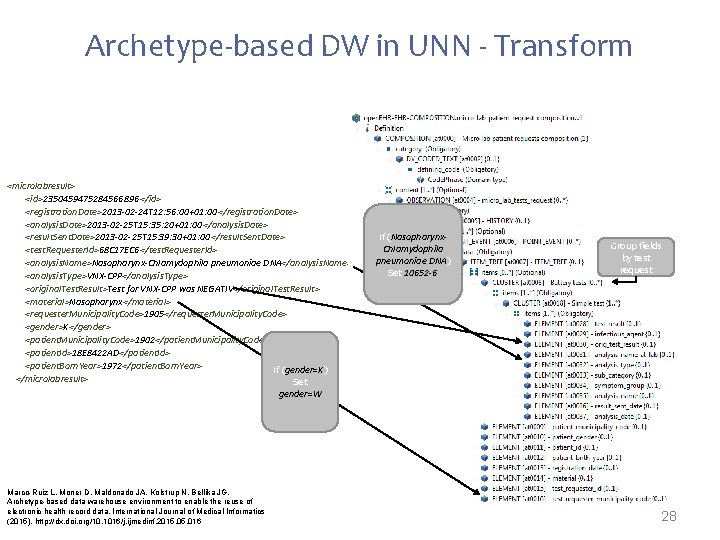
Archetype-based DW in UNN - Transform <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Nasopharynx-Chlamydophila pneumoniae DNA</analysis. Name> <analysis. Type>VNX-CPP</analysis. Type> <original. Test. Result>Test for VNX-CPP was NEGATIV</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>18 E 8422 AD</patient. Id> <patient. Born. Year>1972</patient. Born. Year> If (gender=K) </microlabresult> Set If (Nasopharynx. Chlamydophila pneumoniae DNA) Set 10652 -6 Group fields by test request gender=W Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 28
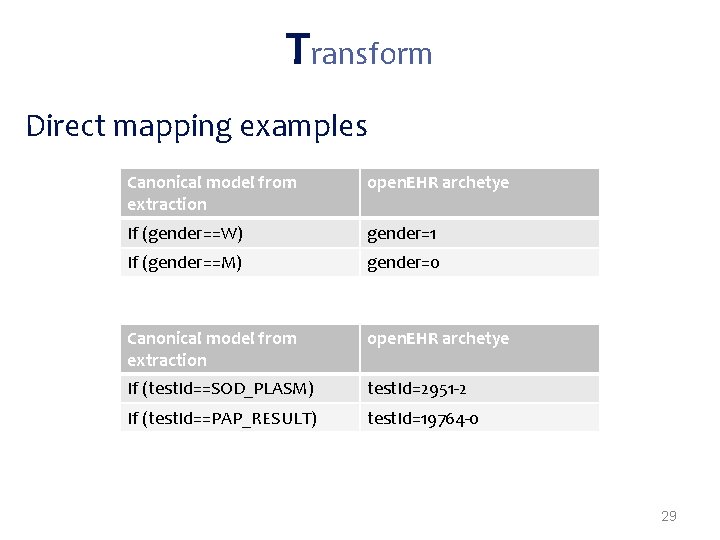
Transform Direct mapping examples Canonical model from extraction open. EHR archetye If (gender==W) gender=1 If (gender==M) gender=0 Canonical model from extraction open. EHR archetye If (test. Id==SOD_PLASM) test. Id=2951 -2 If (test. Id==PAP_RESULT) test. Id=19764 -0 29
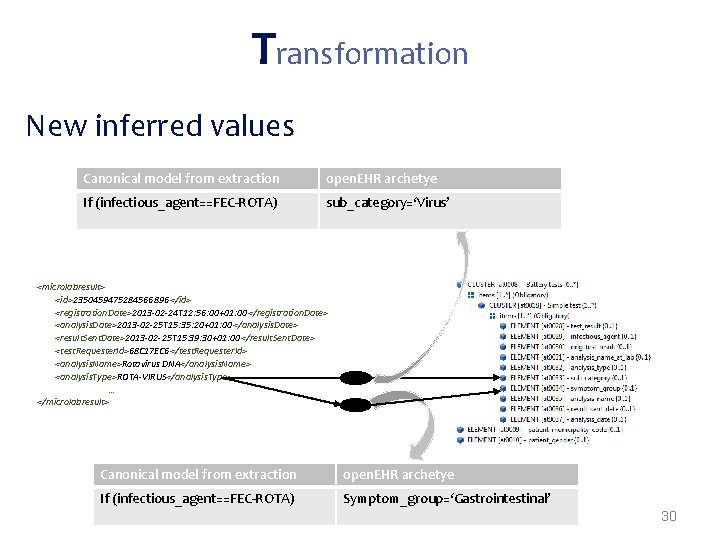
Transformation New inferred values Canonical model from extraction open. EHR archetye If (infectious_agent==FEC-ROTA) sub_category=‘Virus’ <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Rotavirus DNA</analysis. Name> <analysis. Type>ROTA-VIRUS</analysis. Type> … </microlabresult> Canonical model from extraction open. EHR archetye If (infectious_agent==FEC-ROTA) Symptom_group=‘Gastrointestinal’ 30
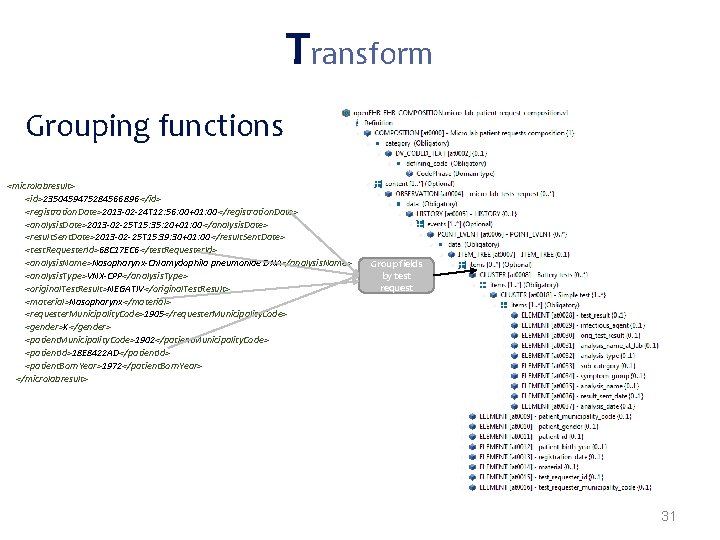
Transform Grouping functions <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Nasopharynx-Chlamydophila pneumoniae DNA</analysis. Name> <analysis. Type>VNX-CPP</analysis. Type> <original. Test. Result>NEGATIV</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>18 E 8422 AD</patient. Id> <patient. Born. Year>1972</patient. Born. Year> </microlabresult> Group fields by test request 31

Extract and Transform techs • Technologies available to extract and transform: – Link. EHR (archetype-based) Transform - Commercial – Pentaho Data Integration (Kettle) ETL - Open Source – Altova Mapforce (Mapping between models) ETL Commercial – Informatica - Commercial – … – Ad-hoc solutions (e. g. java + Drools) • Load needs to be ad-hoc: no commercial open. EHR connectors available 32
Load After transformation, open. EHR extracts are interoperable with other open. EHR systems However, appropriate query mechanisms based on archetypes need to guarantee open. EHR extracts availability and appropriate response times Performing transformations on demand would not ensure efficient responses neither allow the appropriate filtering An open. EHR persistence platform needs to be loaded to enable queries Such platform will allow the retrieval of the standard extracts any time with AQL 33
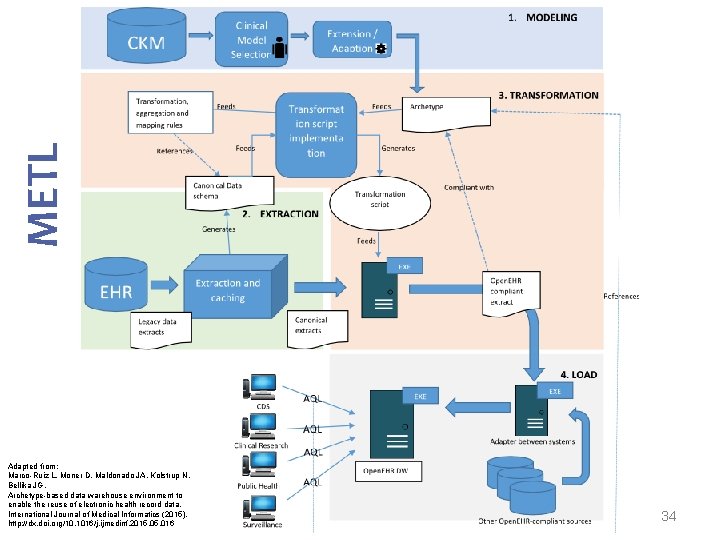
METL Adapted from: Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 34
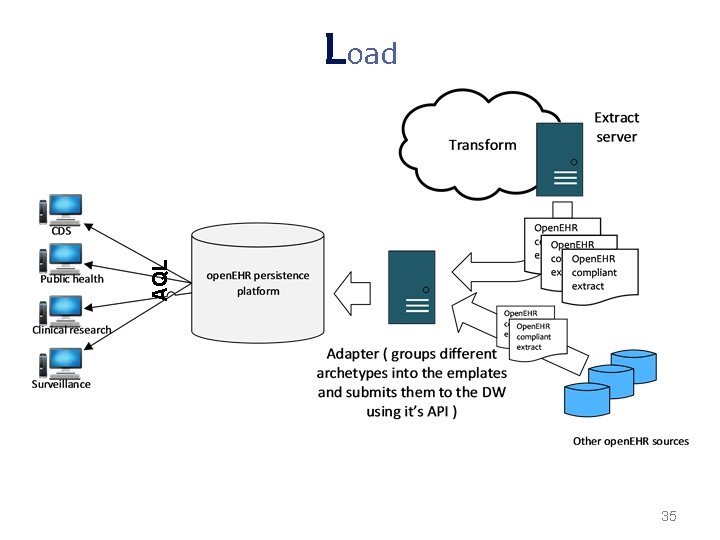
AQL Load 35
Load Reconciliation of formats unveils the need a connectathon to test real Si. Op Load processes are long lasting (hours) Load should be implemented as batch scheduled tasks that do not interfere the query load of the DW 36
Load Ideally the open. EHR EXTRACT IM should be used to encapsulate compositions. This guarantees appropriate version control for data updates However, the EXTRACT model has not catch on in industrial implementations and direct COMPOSITION serializations are used Since data sources are not open. EHR systems, even with the EXTRACT IM, versioning control would present challenges Data updates of the DW must be carefully performed 37

Load The load process can be treated as a transaction global Global transactions not properly managed may incur in wrong inferences when querying the DW The control of data validity across the whole pipeline is still an open issue 38
METL Adapted from: Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 39
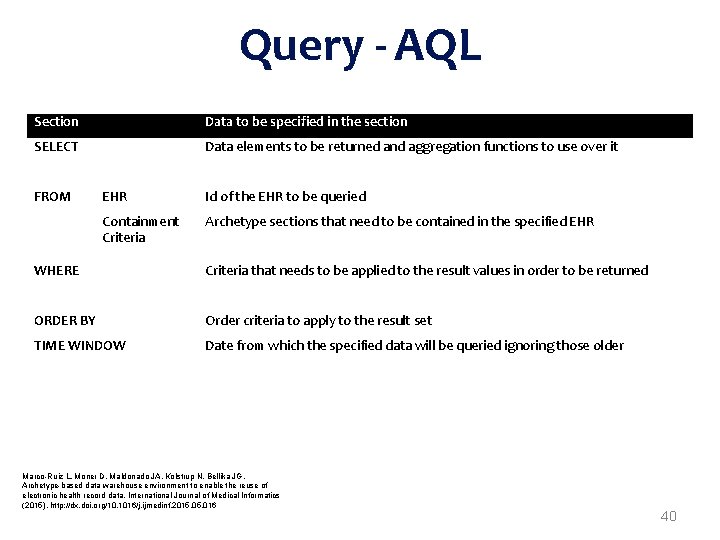
Query - AQL Section Data to be specified in the section SELECT Data elements to be returned and aggregation functions to use over it FROM EHR Id of the EHR to be queried Containment Criteria Archetype sections that need to be contained in the specified EHR WHERE Criteria that needs to be applied to the result values in order to be returned ORDER BY Order criteria to apply to the result set TIME WINDOW Date from which the specified data will be queried ignoring those older Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 40
![AQL - Query samples SELECT o/data/events/data/items[at 0078. 13]/value AS White. Cell. Count FROM EHR[ehr_id=1 AQL - Query samples SELECT o/data/events/data/items[at 0078. 13]/value AS White. Cell. Count FROM EHR[ehr_id=1](http://slidetodoc.com/presentation_image_h2/9daf42bec179b7c34b971a50b4baa298/image-41.jpg)
AQL - Query samples SELECT o/data/events/data/items[at 0078. 13]/value AS White. Cell. Count FROM EHR[ehr_id=1 ADC 27] CONTAINS COMPOSITION c [open. EHR-EHRCOMPOSITION. encounter. v 1] CONTAINS OBSERVATION o [open. EHR-EHROBSERVATION. lab_test_full_blood_count. v 1] WHERE o/data/events/data/items[at 0078. 13]/value > 1100000 AND o/data/events/data/items[at 0078. 13]/value < 1700000 TIME WINDOW P 1 Y/2014 -02 -12 Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 41
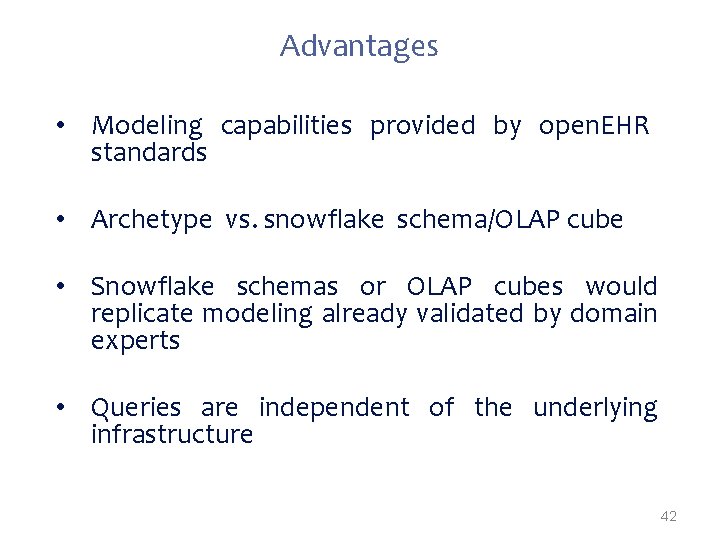
Advantages • Modeling capabilities provided by open. EHR standards • Archetype vs. snowflake schema/OLAP cube • Snowflake schemas or OLAP cubes would replicate modeling already validated by domain experts • Queries are independent of the underlying infrastructure 42

Limitations • Limited control over ETL stages. Global transactions need to be implemented. • Synchronization and version control issues can arise when integrating several sources and deciding which entities need to be updated • Load not rolled back will lead to wrong inferences • Rules involving time cannot be easily implemented 43
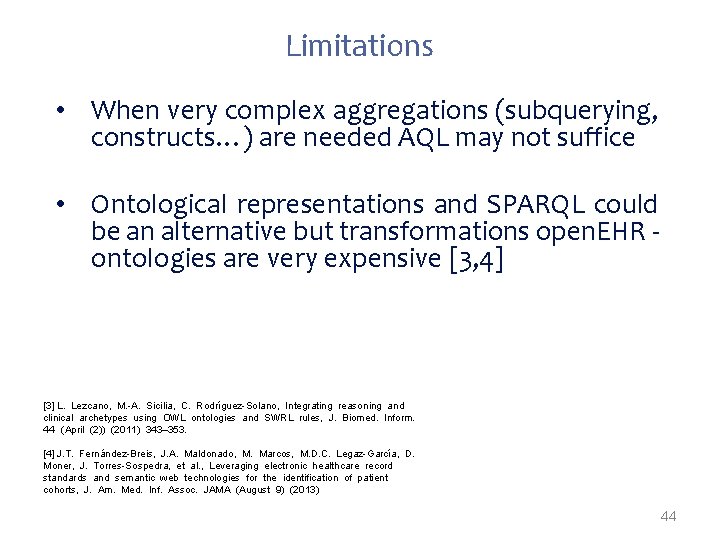
Limitations • When very complex aggregations (subquerying, constructs…) are needed AQL may not suffice • Ontological representations and SPARQL could be an alternative but transformations open. EHR ontologies are very expensive [3, 4] [3] L. Lezcano, M. -A. Sicilia, C. Rodríguez-Solano, Integrating reasoning and clinical archetypes using OWL ontologies and SWRL rules, J. Biomed. Inform. 44 (April (2)) (2011) 343– 353. [4] J. T. Fernández-Breis, J. A. Maldonado, M. Marcos, M. D. C. Legaz-García, D. Moner, J. Torres-Sospedra, et al. , Leveraging electronic healthcare record standards and semantic web technologies for the identification of patient cohorts, J. Am. Med. Inf. Assoc. JAMA (August 9) (2013) 44

Agenda 1. Background 1. 1. Learning Healthcare System 1. 2. Semantic Interoperability 1. 3. Linkage EHR – Inference models 2. METL 2. 1. Modelling 2. 2. Extract 2. 3. Transform 2. 4. Load 3. Use cases 3. 1. Laboratory Service at University Hospital North Norway 3. 2. NZ Cardiac Registry 3. 3. Path based queries 45

Use cases

Infectious diseases tests monitoring at University Hospital of North Norway

Archetype-based DW at UNN - Introduction • Population information for general practitioners (GPs) is usually limited by the patients they are assigned and their personal communications with colleagues • They seldom have access to real time population test results or colleagues requests • Access to anonymized and aggregated population data about laboratory interventions of other colleagues and laboratory personnel can empower their environmental awareness of communicable infectious diseases and help them to determine which set of tests should be ordered 48
Archetype-based DW at UNN - Introduction • Laboratory test results of a population of 230, 000 patients belonging to Troms and Finnmark counties in Norway requested between January 2013 and November 2014 were normalized to open. EHR • Test records normalization has been performed by defining transformation and aggregation functions to automatically generate open. EHR compliant data. • These data were loaded into an archetype-based data warehouse 49

Archetype-based DW in UNN - Introduction • Indicators linked to the data in the warehouse to monitor test activity of Salmonella and Pertussis were defined with AQL 50
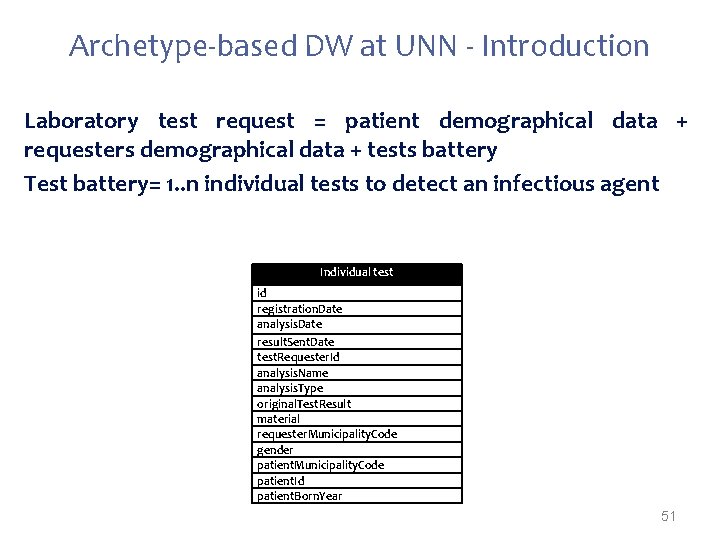
Archetype-based DW at UNN - Introduction Laboratory test request = patient demographical data + requesters demographical data + tests battery Test battery= 1. . n individual tests to detect an infectious agent Individual test id registration. Date analysis. Date result. Sent. Date test. Requester. Id analysis. Name analysis. Type original. Test. Result material requester. Municipality. Code gender patient. Municipality. Code patient. Id patient. Born. Year 51
Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 52

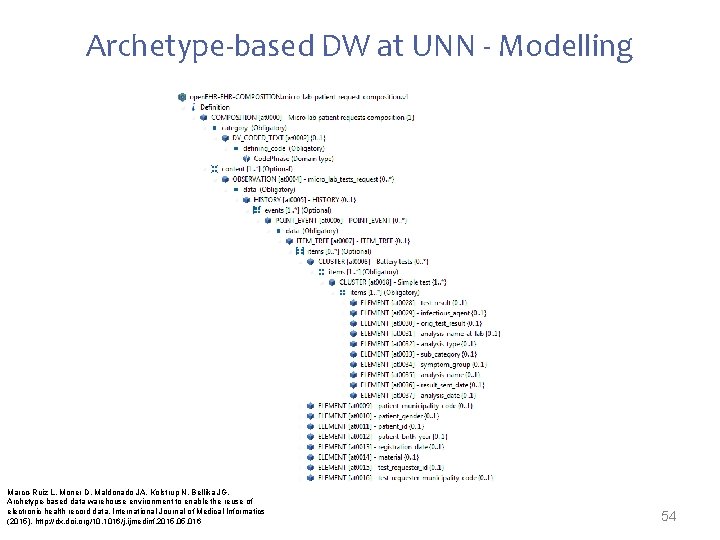
Archetype-based DW at UNN - Modelling • Reuse was attempted checking the international open. EHR CKM • 2 possible candidates were identified – open. EHR-OBSERVATION. lab _test. v 1 – open. EHR-OBSERVATION. lab test-microbiology. v 1 Specialize • The need of demographical information and fields like infectious agent or symptom group forced the definition of new archetypes 53
Archetype-based DW at UNN - Modelling Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 54
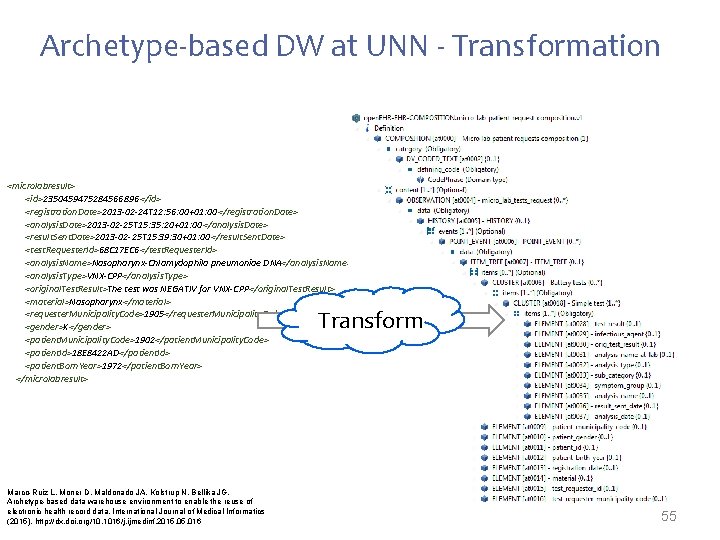
Archetype-based DW at UNN - Transformation <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Nasopharynx-Chlamydophila pneumoniae DNA</analysis. Name> <analysis. Type>VNX-CPP</analysis. Type> <original. Test. Result>The test was NEGATIV for VNX-CPP</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>18 E 8422 AD</patient. Id> <patient. Born. Year>1972</patient. Born. Year> </microlabresult> Transform Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 55
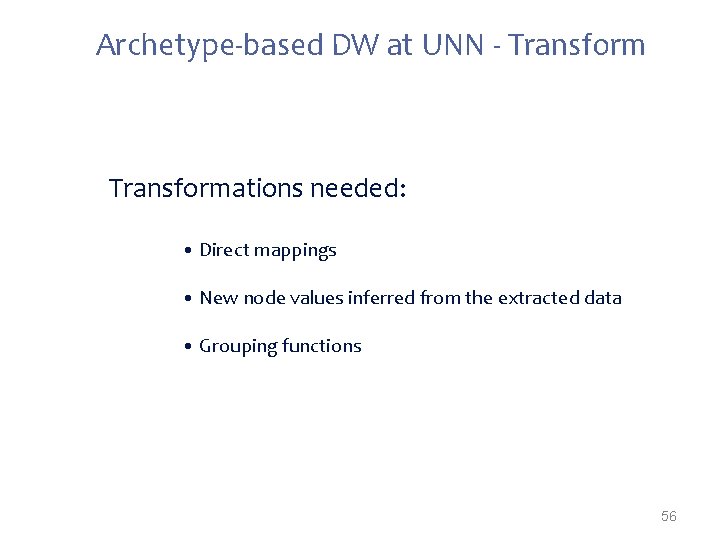
Archetype-based DW at UNN - Transformations needed: • Direct mappings • New node values inferred from the extracted data • Grouping functions 56
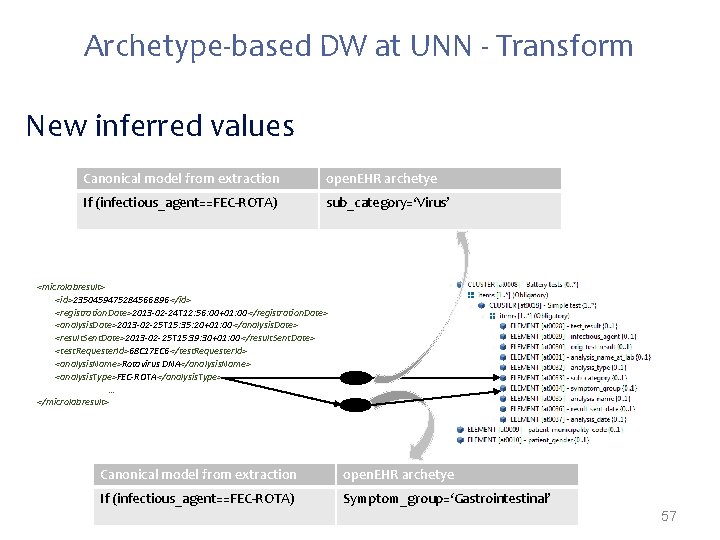
Archetype-based DW at UNN - Transform New inferred values Canonical model from extraction open. EHR archetye If (infectious_agent==FEC-ROTA) sub_category=‘Virus’ <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Rotavirus DNA</analysis. Name> <analysis. Type>FEC-ROTA</analysis. Type> … </microlabresult> Canonical model from extraction open. EHR archetye If (infectious_agent==FEC-ROTA) Symptom_group=‘Gastrointestinal’ 57
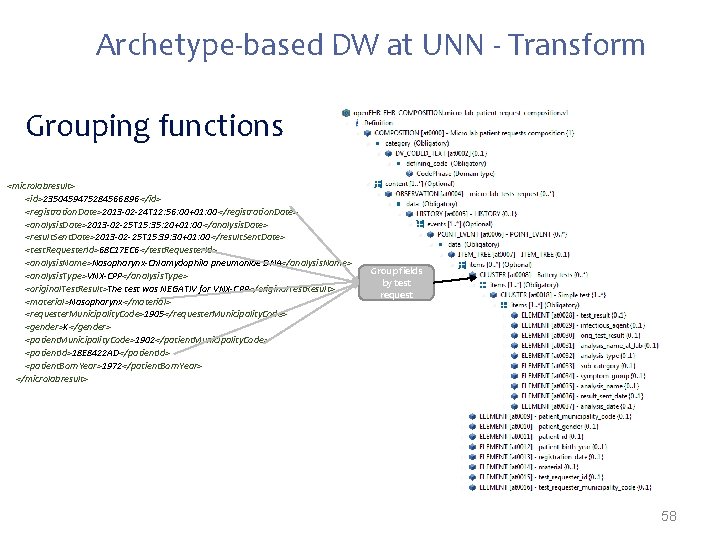
Archetype-based DW at UNN - Transform Grouping functions <microlabresult> <id>2350459475284566896 </id> <registration. Date>2013 -02 -24 T 12: 56: 00+01: 00</registration. Date> <analysis. Date>2013 -02 -25 T 15: 35: 20+01: 00</analysis. Date> <result. Sent. Date>2013 -02 -25 T 15: 39: 30+01: 00</result. Sent. Date> <test. Requester. Id>68 C 17 EC 6</test. Requester. Id> <analysis. Name>Nasopharynx-Chlamydophila pneumoniae DNA</analysis. Name> <analysis. Type>VNX-CPP</analysis. Type> <original. Test. Result>The test was NEGATIV for VNX-CPP</original. Test. Result> <material>Nasopharynx</material> <requester. Municipality. Code>1905</requester. Municipality. Code> <gender>K</gender> <patient. Municipality. Code>1902</patient. Municipality. Code> <patient. Id>18 E 8422 AD</patient. Id> <patient. Born. Year>1972</patient. Born. Year> </microlabresult> Group fields by test request 58
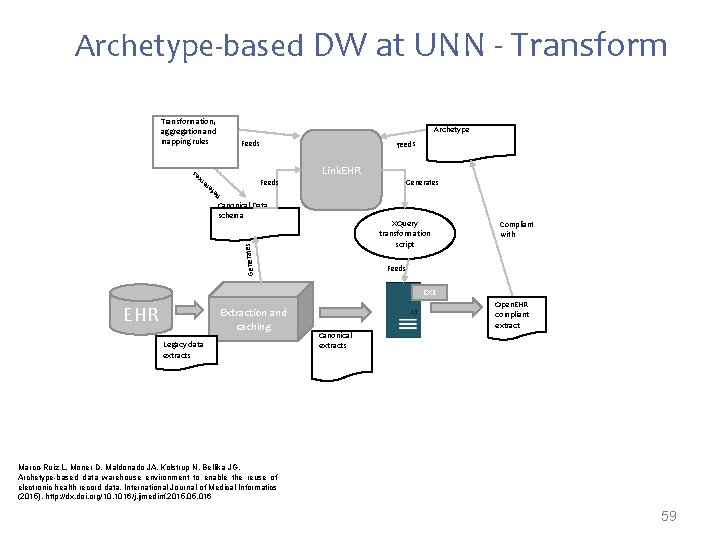
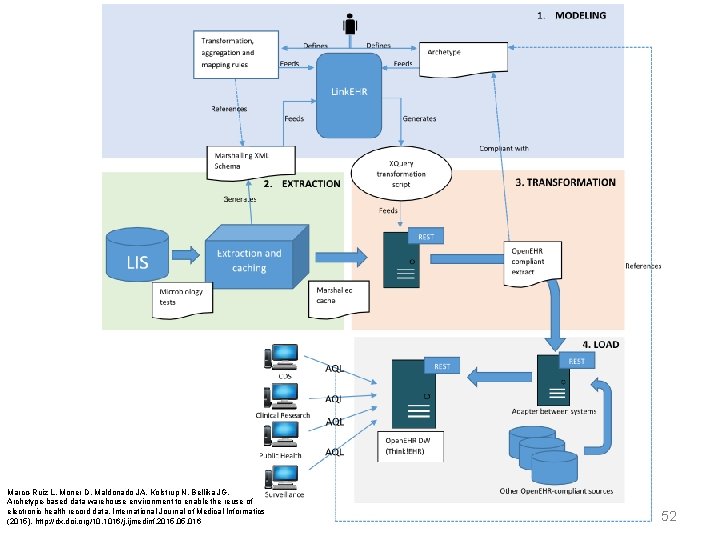
Archetype-based DW at UNN - Transformation, aggregation and mapping rules Archetype Feeds Link. EHR Feeds Generates Re f es nc er e Canonical Data schema Generates XQuery transformation script Compliant with Feeds EXE EHR Extraction and caching Legacy data extracts Open. EHR compliant extract Canonical extracts Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 59

Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 Transformation 60
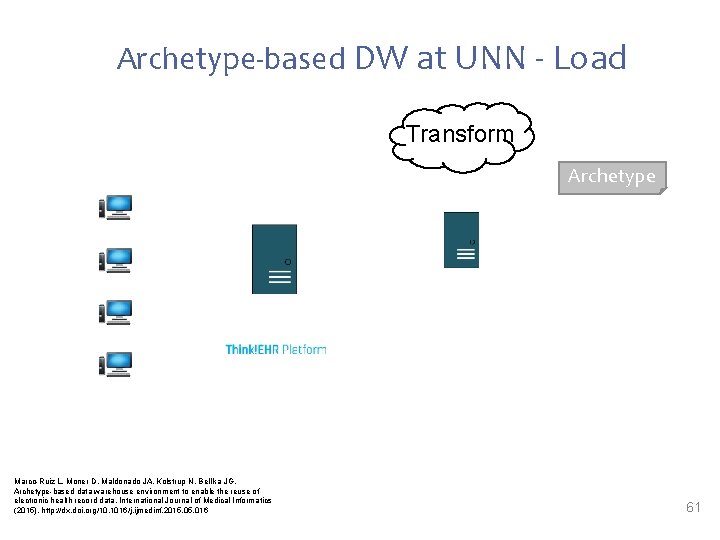
Archetype-based DW at UNN - Load Transform Archetype Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 61

Archetype-based DW at UNN - Load • Load was performed sequentially for each patient calling the extracts service from the Transform stage • The extracts where simple COMPOSITION serializations. The EXTRACT IM has not been used • Some differences between the seriations from the Transformation stage and the serializations accepted by the DW were found (namespaces, message wrapping…) • Format reconciliation was needed • The open. EHR project Connectathon should guarantee open. EHR tooling to interoperate seamlessly • We defined several indicators to monitor infectious diseases (salmonella and pertussis) 62

Archetype-based DW at UNN - Query • After loading the DW we were able to query data creating different data sets for different scenarios • As use case we defined several indicators to monitor infectious diseases (salmonella and pertussis) 63
Archetype-based DW at UNN Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 64
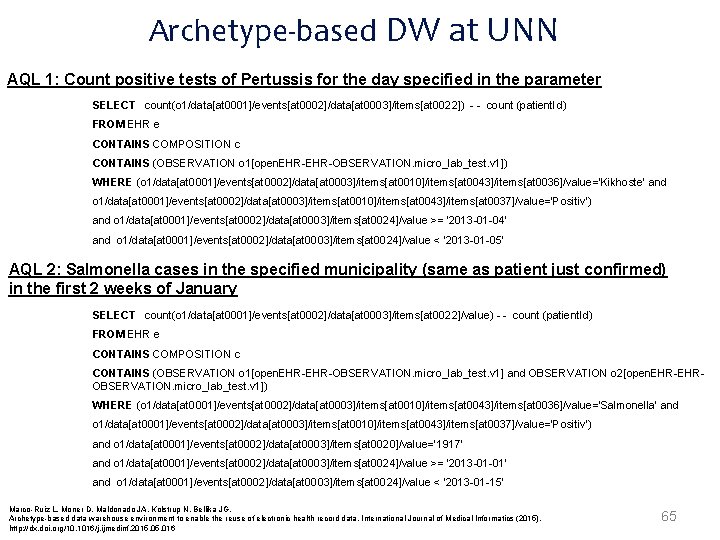
Archetype-based DW at UNN AQL 1: Count positive tests of Pertussis for the day specified in the parameter SELECT count(o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0022]) - - count (patient. Id) FROM EHR e CONTAINS COMPOSITION c CONTAINS (OBSERVATION o 1[open. EHR-OBSERVATION. micro_lab_test. v 1]) WHERE (o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0010]/items[at 0043]/items[at 0036]/value='Kikhoste‘ and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0010]/items[at 0043]/items[at 0037]/value='Positiv') and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0024]/value >= '2013 -01 -04' and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0024]/value < '2013 -01 -05' AQL 2: Salmonella cases in the specified municipality (same as patient just confirmed) in the first 2 weeks of January SELECT count(o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0022]/value) - - count (patient. Id) FROM EHR e CONTAINS COMPOSITION c CONTAINS (OBSERVATION o 1[open. EHR-OBSERVATION. micro_lab_test. v 1] and OBSERVATION o 2[open. EHR-EHROBSERVATION. micro_lab_test. v 1]) WHERE (o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0010]/items[at 0043]/items[at 0036]/value='Salmonella‘ and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0010]/items[at 0043]/items[at 0037]/value='Positiv') and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0020]/value='1917' and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0024]/value >= '2013 -01 -01' and o 1/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0024]/value < '2013 -01 -15' Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 65

Archetype-based DW at UNN Work available at: Marco-Ruiz L, Moner D, Maldonado JA, Kolstrup N, Bellika JG, Archetype-based data warehouse environment to enable the reuse of electronic health record data, International Journal of Medical Informatics (2015), http: //dx. doi. org/10. 1016/j. ijmedinf. 2015. 016 Special thanks to: This work was supported by Helse Nord [grant HST 1121 -13 and 9057/HST 1120 -13]; the NILS Science and Sustainability Programme [grant number 005 -ABEL-IM-2013] from Iceland, Liechtenstein and Norway through the EEA Financial Mechanism, operated by Universidad Complutense de Madrid; and by the Spanish Ministry of Economy and Competitiveness [grant PTQ-12 -05620]. We would like to thank to Marand d. o. o. and Torje S. Henriksen for the products provided and their assistance and support during this work. We would like to acknowledge Gunnar Skov Simonsen and Marit Wiklund at the microbiology laboratory service of the University Hospital of North Norway for their support for this work. 66

Recent developments and future plans • • Additional microbiology labs have joined Snow Complete coverage of Northern Norway Soon partly coverage of whole Norway Tasks involved in setting up a new laboratory: – – – Establishing network connection Setting up a physical / virtual Snow Server Defining laboratory analysis code mapping rules Initiating data extracts Defining data import transformations Setting up Snow data consumption missions for epidemiology model generation – Preparing visualisation of epidemiology model data 67
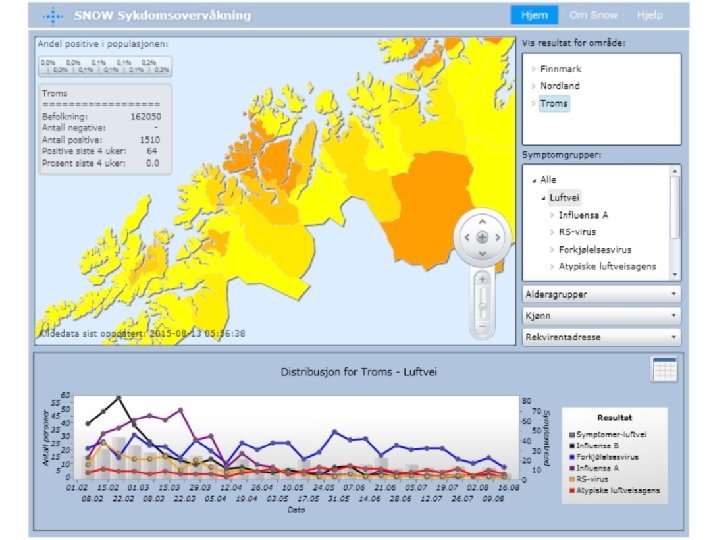
68
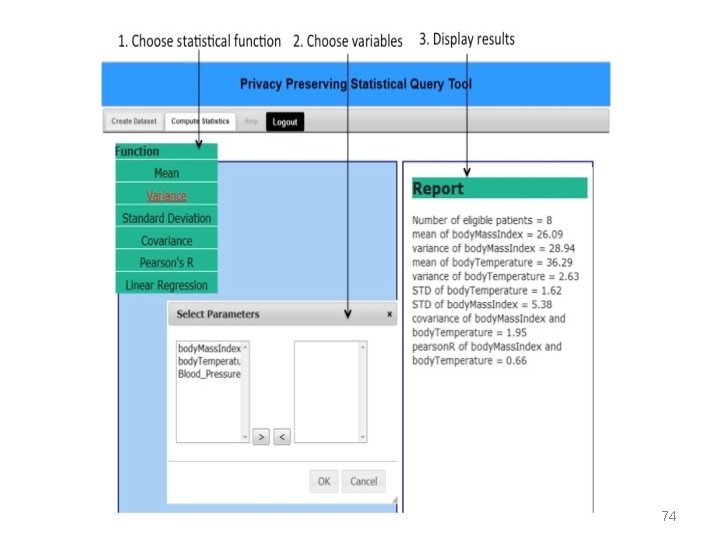
Future plans – distributed analysis of Open. EHR data using secure multiparty computations • A two-phase solution 1. Dataset creation 2. Statistical computation More details available in: • M. A. Hailemichael, L. Marco-Ruiz, and J. G. Bellika, “Privacy-preserving Statistical Query and Processing on Distributed Open. EHR Data, ” Stud Health Technol Inform, vol. 210, pp. 766– 770, 2015. • Meskerem Asfaw Hailemichael, Kassaye Yitbarek Yigzaw, Johan Gustav Bellika (2015). Emnet: a System for Privacy-Preserving Statistical Computing on Distributed Health Data, SHI 2015, Proceedings from The 13 th Scandinavien Conference on Health Informatics, June 15– 17, 2015, Tromsø, Norway 27. 05. 15 http: //www. ep. liu. se/ecp_article/index. en. aspx? issue=115; article=006 69

Open. EHR in Norway • The current strategic plan of Norwegian Health authorities is encouraging EHR vendors to adopt open. EHR 1 – DIPS ASA, which is the provider of more than 70% of hospital EHRs in Norway, is using Open. EHR 2 • Norwegian CKM 1 Ellingsen G, Christensen B, Silsand L. Developing Large-scale Electronic Patient Records Conforming to the open. EHR Architecture. Procedia Technology. 2014; 16: 1281– 6. “ 2 http: //www. dips. no/eng/about-us/customers? lang=eng 27. 05. 15 70
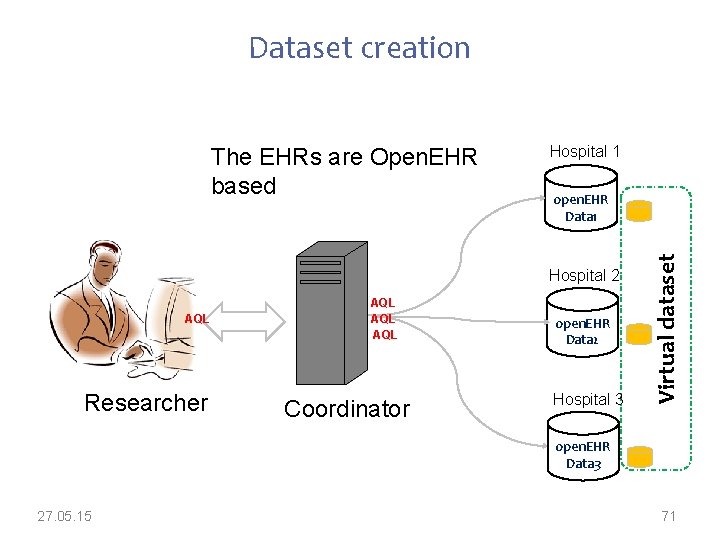
Dataset creation Hospital 1 open. EHR Data 1 Hospital 2 AQL Researcher AQL AQL Coordinator open. EHR Data 2 Hospital 3 Virtual dataset The EHRs are Open. EHR based open. EHR Data 3 27. 05. 15 71
Web-klient 72
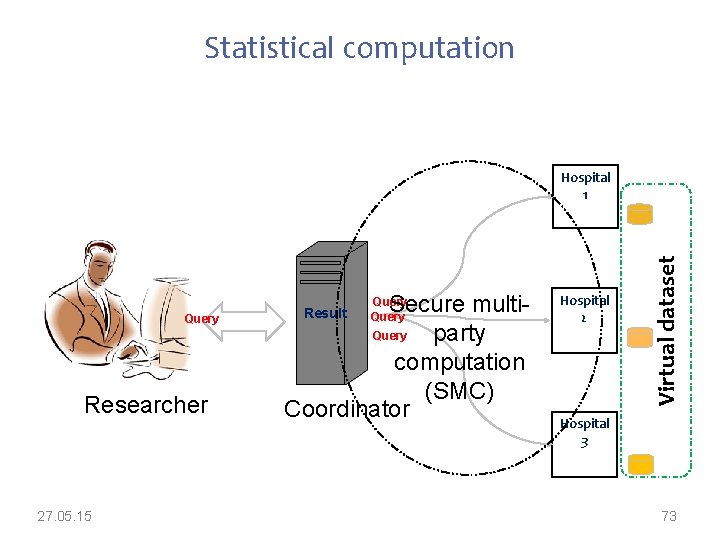
Statistical computation Query Researcher 27. 05. 15 Secure multi. Query party computation (SMC) Coordinator Result Query Hospital 2 Virtual dataset Hospital 1 Hospital 3 73
74
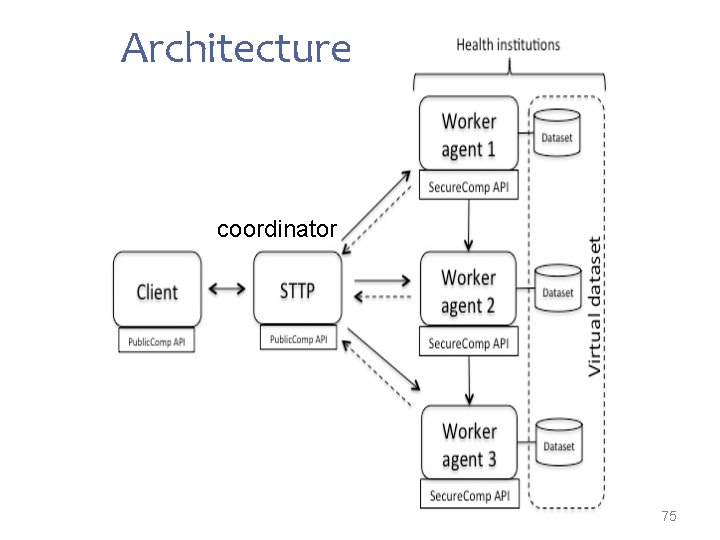
Architecture coordinator 75

Relevant publications • • • L. Marco-Ruiz, D. Moner, J. A. Maldonado, N. Kolstrup, and J. G. Bellika, “Archetype-based data warehouse environment to enable the reuse of electronic health record data, ” International Journal of Medical Informatics, vol. 84, no. 9, pp. 702– 714, Sep. 2015. J. G. Bellika, T. Henriksen, and K. Y. Yigzaw, “The Snow System – A Decentralized Medical Data Processing System, ” in Data Mining in Clinical Medicine, vol. 1246, Spinger, 2014. M. A. Hailemichael, L. Marco-Ruiz, and J. G. Bellika, “Privacy-preserving Statistical Query and Processing on Distributed Open. EHR Data, ” Stud Health Technol Inform, vol. 210, pp. 766– 770, 2015. Meskerem Asfaw Hailemichael, Kassaye Yitbarek Yigzaw, Johan Gustav Bellika (2015). Emnet: a System for Privacy-Preserving Statistical Computing on Distributed Health Data, SHI 2015, Proceedings from The 13 th Scandinavien Conference on Health Informatics, June 15– 17, 2015, Tromsø, Norway http: //www. ep. liu. se/ecp_article/index. en. aspx? issue=115; article=006 (accessed 8/18/2015) J. G. Bellika, T. Hasvold, and G. Hartvigsen, “Propagation of program control: a tool for distributed disease surveillance, ” Int J Med Inform, vol. 76, no. 4, pp. 313– 29, 2007. J. G. Bellika, H. Sue, L. Bird, A. Goodchild, T. Hasvold, and G. Hartvigsen, “Properties of a federated epidemiology query system, ” Int J Med Inform, vol. 76, no. 9, pp. 664– 76, 2007. 76

Agenda 1. Background 1. 1. Learning Healthcare System 1. 2. Semantic Interoperability 1. 3. Linkage EHR – Inference models 2. METL 2. 1. Modelling 2. 2. Extract 2. 3. Transform 2. 4. Load 3. Use cases 3. 1. Laboratory Service at University Hospital North Norway 3. 2. NZ Cardiac Registry 3. 3. Path based queries 77
ANZACS-QI* open. EHR Modelling for Datawarehousing Koray Atalag Jane Farris *All NZ Acute Coronary Syndrome Quality Improvement programme National clinical registry for acute coronary syndrome (ACS) events and cardiac procedures
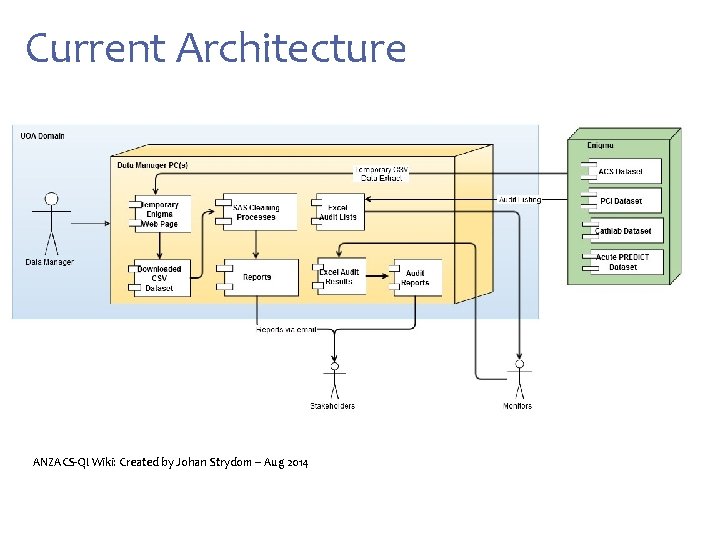
Current Architecture ANZACS-QI Wiki: Created by Johan Strydom – Aug 2014

Current Situation • Flat files transferred from Enigma • Heavily dependent on Data Dictionary for meaning (Word & Excel files) • No view ‘across’ datasets • Requirement for extensive clinical input for report development and on-going support
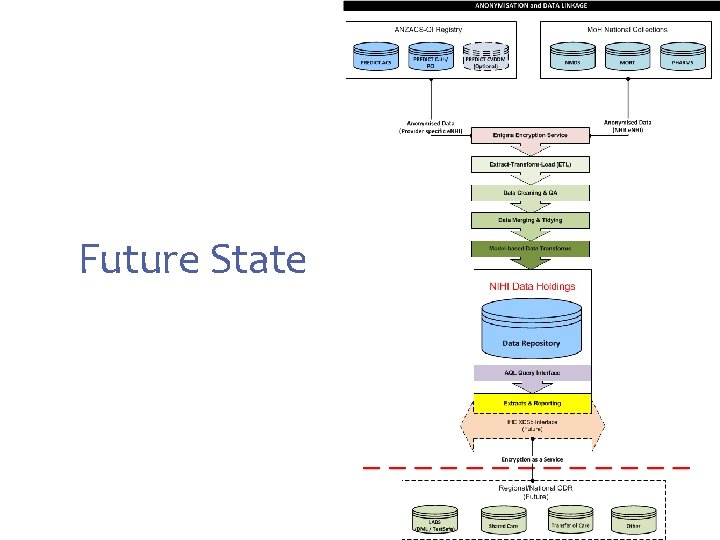
Future State

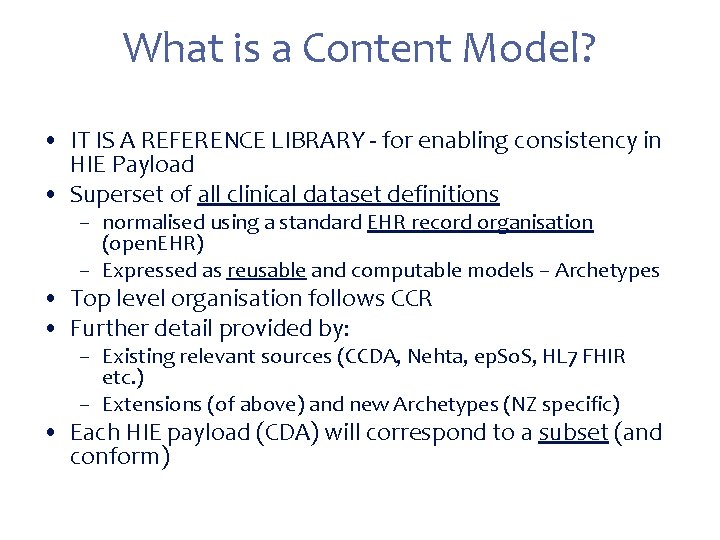
What is a Content Model? • IT IS A REFERENCE LIBRARY - for enabling consistency in HIE Payload • Superset of all clinical dataset definitions – normalised using a standard EHR record organisation (open. EHR) – Expressed as reusable and computable models – Archetypes • Top level organisation follows CCR • Further detail provided by: – Existing relevant sources (CCDA, Nehta, ep. So. S, HL 7 FHIR etc. ) – Extensions (of above) and new Archetypes (NZ specific) • Each HIE payload (CDA) will correspond to a subset (and conform)
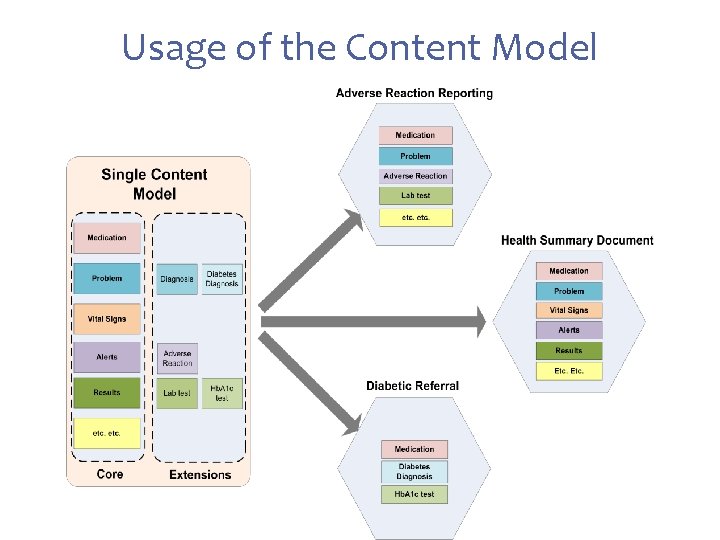
Usage of the Content Model
Exploiting Content Model for Secondary Use Atalag K. Using a single content model for e. Health interoperability and secondary use. Stud Health Technol Inform. 2013; 193: 282– 96
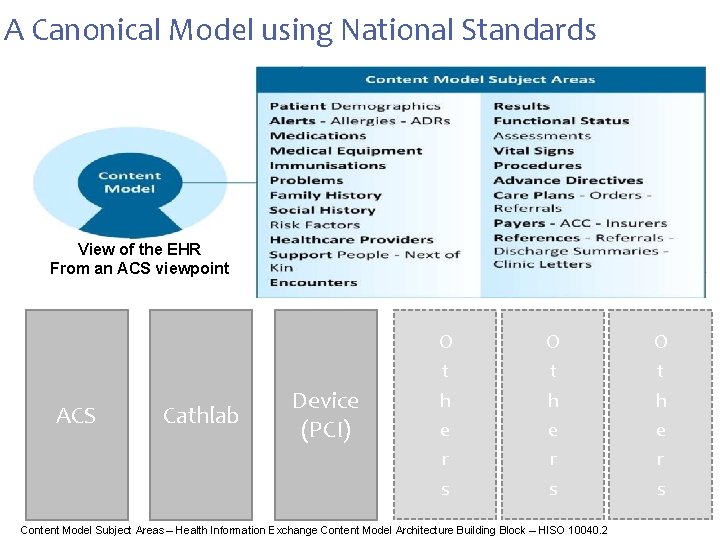
A Canonical Model using National Standards View of the EHR From an ACS viewpoint ACS Cathlab Device (PCI) O t h e r s Content Model Subject Areas – Health Information Exchange Content Model Architecture Building Block – HISO 10040. 2 O t h e r s
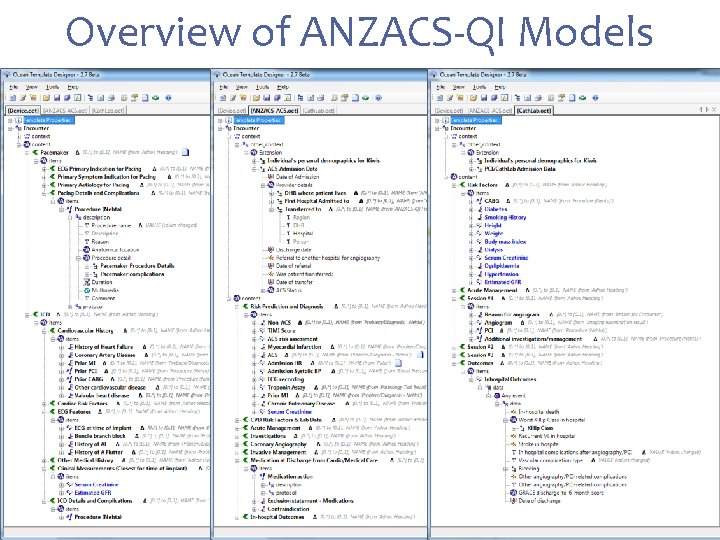
Overview of ANZACS-QI Models
Benefits • Single point of reference – Faithful representation of the ‘forms’ – Standards based • Extensible • Flexible • Reusable – Clear and unambiguous data definition – Enables single source metadata management • Data Dictionary • Rules within and between forms • Rules to other data sources (e. g. Linking datasets) – Export for reuse – Holds the clinical viewpoint – ACS part of the EHR
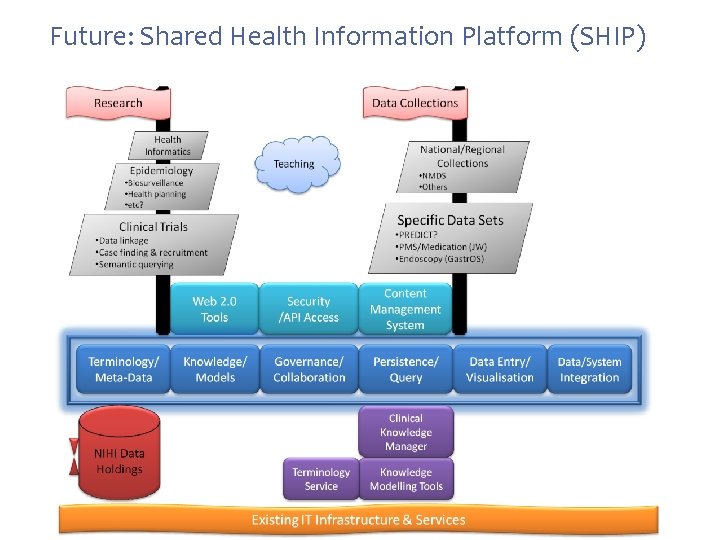
Future: Shared Health Information Platform (SHIP)

Agenda 1. Background 1. 1. Learning Healthcare System 1. 2. Semantic Interoperability 1. 3. Linkage EHR – Inference models 2. METL 2. 1. Modelling 2. 2. Extract 2. 3. Transform 2. 4. Load 3. Use cases 3. 1. Laboratory Service at University Hospital North Norway 3. 2. NZ Cardiac Registry 3. 3. Path based queries 90

Path-based queries in action Test available in EHRServer https: //cabolabs-ehrserver. rhcloud. com/ehr-0. 3/query/list
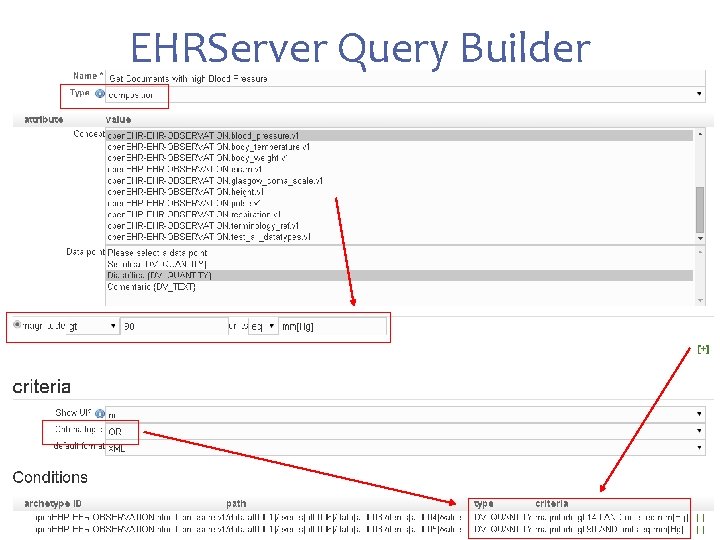
EHRServer Query Builder

Path-based queries in action Path-based: + Get clinical documents (compositions) + With high BP { JSON expression of EHRServer queries "uid": "9 c 5 da 334 -4 b 81 -4 d 60 -92 e 2 -aa 96 a 722 b 4 ac" , "name": "Documents with high BP", "format": "xml", "type": "composition", "criteria. Logic": "OR", "criteria": [ { "archetype. Id": "open. EHR-OBSERVATION. blood_pressure. v 1", "path": "/data[at 0001]/events[at 0006]/data[at 0003]/items[at 0004]/value" , "conditions": { "magnitude": { "gt": [ 140 ] }, "units": { "eq": "mm[Hg]" } } }, { "archetype. Id": "open. EHR-OBSERVATION. blood_pressure. v 1", "path": "/data[at 0001]/events[at 0006]/data[at 0003]/items[at 0005]/value" , "conditions": { "magnitude": { "gt": [ 90 ] }, "units": { "eq": "mm[Hg]" } } } ] }

Path-based queries in action Results: + in XML (or JSON if specified on the query or as a parameter) + just the index, no data, can get a specific document using the index <? xml version="1. 0" encoding="UTF-8"? > <list> <composition. Index id="8"> <archetype. Id>open. EHR-COMPOSITION. signos. v 1</archetype. Id> <category>event</category> <data. Indexed>true</data. Indexed> <ehr. Id>1111 -1111 -11111111 </ehr. Id> <last. Version>true</last. Version> <start. Time>2015 -08 -14 03: 06: 44. 0 EDT</start. Time> <subject. Id>1111 -1111 -11111111 </subject. Id> <template. Id>Signos</template. Id> <uid>e 152 b 2 c 2 -7 dbe-44 b 6 -9 ec 6 -2 cd 698561140</uid> </composition. Index> <composition. Index id="9"> <archetype. Id>open. EHR-COMPOSITION. signos. v 1</archetype. Id> <category>event</category> <data. Indexed>true</data. Indexed> <ehr. Id>1111 -1111 -11111111 </ehr. Id> <last. Version>true</last. Version> <start. Time>2015 -08 -14 03: 07: 06. 0 EDT</start. Time> <subject. Id>1111 -1111 -11111111 </subject. Id> <template. Id>Signos</template. Id> <uid>f 0 a 8 d 192 -0 f 68 -4501 -8373 -f 954 a 47 a 7385</uid> </composition. Index>. . . </list>

Path-based queries in action Path-based: + Get clinical data for all vital signs measures + Result in JSON format, grouped by path (type of data) { "uid": "70764 d 85 -4 e 4 b-4548 -8 f 71 -3 a 294 f 35 e 704" , "name": "Vital Signs", "format": "json", "type": "datavalue", "group": "path", "projections": [ { "archetype. Id": "open. EHR-OBSERVATION. blood_pressure. v 1", "path": "/data[at 0001]/events[at 0006]/data[at 0003]/items[at 0004]/value" }, { "archetype. Id": "open. EHR-OBSERVATION. blood_pressure. v 1", "path": "/data[at 0001]/events[at 0006]/data[at 0003]/items[at 0005]/value" }, { "archetype. Id": "open. EHR-OBSERVATION. body_temperature. v 1", "path": "/data[at 0002]/events[at 0003]/data[at 0001]/items[at 0004]/value" }, { "archetype. Id": "open. EHR-OBSERVATION. body_weight. v 1", "path": "/data[at 0002]/events[at 0003]/data[at 0001]/items[at 0004]/value" }, { "archetype. Id": "open. EHR-OBSERVATION. pulse. v 1", "path": "/data[at 0002]/events[at 0003]/data[at 0001]/items[at 0004]/value" }, { "archetype. Id": "open. EHR-OBSERVATION. respiration. v 1", "path": "/data[at 0001]/events[at 0002]/data[at 0003]/items[at 0004]/value" } ] } JSON expression of EHRServer queries

Q&A Contact: Luis Marco Luis. Marco. Ruiz@telemed. no Pablo Pazos pablo. pazos@cabolabs. com, Koray Atalag k. atalag@auckland. ac. nz, Johan G. Bellika johan. Gustav. Bellika@telemed. no, Kassaye Y. Yigzaw kassaye. y. yigzaw@uit. no
- Slides: 96