ENABLING ACCESSIBLE AND SENSITIVE SEQUENCE PROFILING FOR SYSTEMS

ENABLING ACCESSIBLE AND SENSITIVE SEQUENCE PROFILING FOR SYSTEMS MEDICINE STEPHANIE I. FRALEY, PH. D. ASSISTANT PROFESSOR BIOENGINEERING, UC SAN DIEGO Disclosure: The technology presented is patented, and I am an inventor on the patents.

SYSTEMS MEDICINE Major goals: • Make blood a diagnostic window of health & disease • Provide deep insights into disease mechanisms in patient-specific contexts Mandates: • Develop technologies to explore patient’s data space in new dimensions • Democratize data-generation & analysis tools Leroy Hood & Mauricio Flores, New Biotechnology 29, 2012.
TECHNOLOGY DEVELOPMENT • Need to measure rapidly and often in humans • • Reduced cost, effort, & time to profile Portability for broad access Time-course readouts to understand disease processes Low level regulators critical to biological meta-stability & contribute to diversity 1, 2 • Higher sensitivity and quantitative power than sequencing & microarrays • Many applications have no need to re-sequence • As de-novo sequencing discovers & catalogs all sequence possibilities, profiling can play a larger and larger role 1. M. B. Elowitz, et. al. , Science 297, 2002. 2. J. L. Spudich & D. E. Koshland. Nature 262, 1976.
BACTERIAL INFECTIONS & SEPSIS • • Hourly mortality risk increase Polymicrobial risk higher Incorrect treatment Antibiotic resistance Spread Biothreat Host-Pathogen systemspecific responses
IDEAL PROFILING TECHNOLOGY FOR DYNAMIC SYSTEMS MEDICINE • Broad-based • Identify each individual sequence present sensitively & specifically • Absolute quantitative power • Rapid • User-independent identification • Today’s sequencing • Sensitivity-specificity trade-off • Speed • Other tech’s have similar limitations
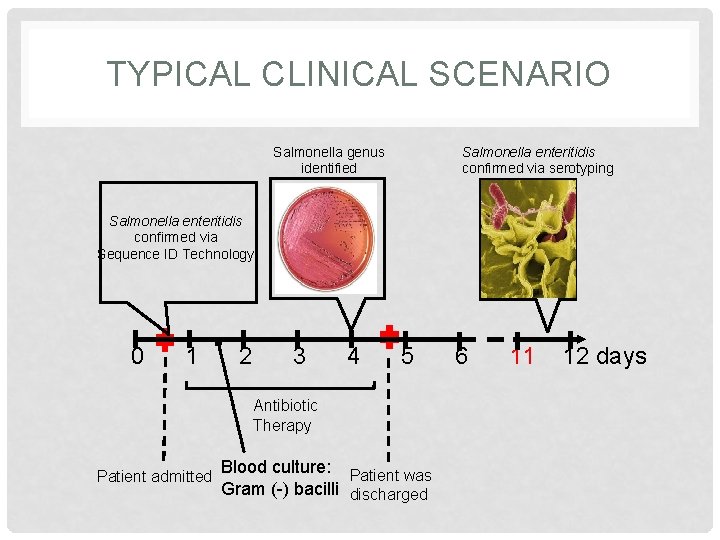
TYPICAL CLINICAL SCENARIO Salmonella genus identified Salmonella enteritidis confirmed via serotyping Salmonella enteritidis confirmed via Sequence ID Technology 0 1 2 3 4 5 Antibiotic Therapy Patient admitted Blood culture: Patient was Gram (-) bacilli discharged 6 11 12 days
OBJECTIVE: PATHOGEN SEQUENCE IDENTIFICATION • In the context of bacterial blood stream infections… • • Broad-based = detect all bacterial pathogens Specific = which pathogens which drugs Sensitive = single cell as dictated by sample limitations Unaffected by contamination Resolve heterogeneity = resolve polymicrobial infections Fast User independent automated analysis
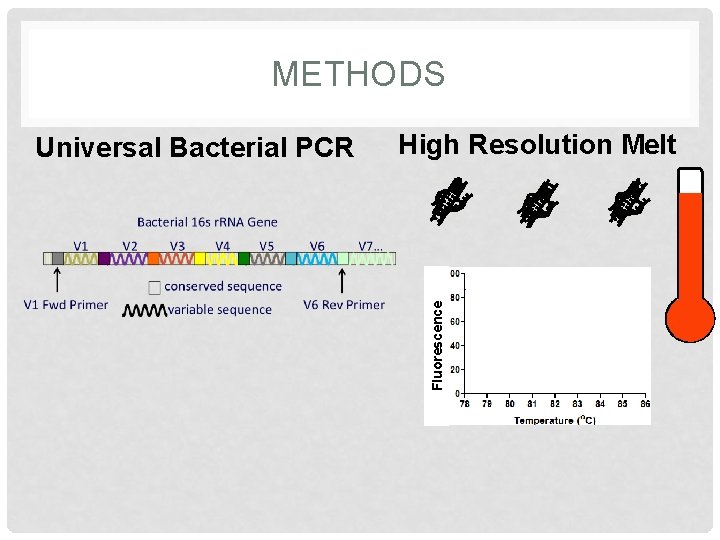
METHODS High Resolution Melt Fluorescence Universal Bacterial PCR
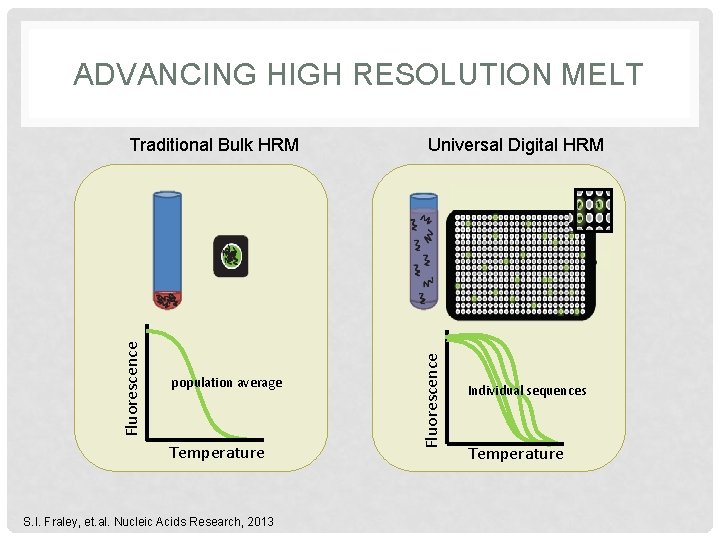
ADVANCING HIGH RESOLUTION MELT population average Temperature S. I. Fraley, et. al. Nucleic Acids Research, 2013 Universal Digital HRM Fluorescence Traditional Bulk HRM Individual sequences Temperature

MICROFLUIDIC DIGITIZATION • Small volume = less reagents, less $$ = greater partitioning of molecules = higher sensitivity • Integrated, automated, disposable – DNA extraction – PCR – Readout • Portable • Rabid heating/cooling ~$15 per test
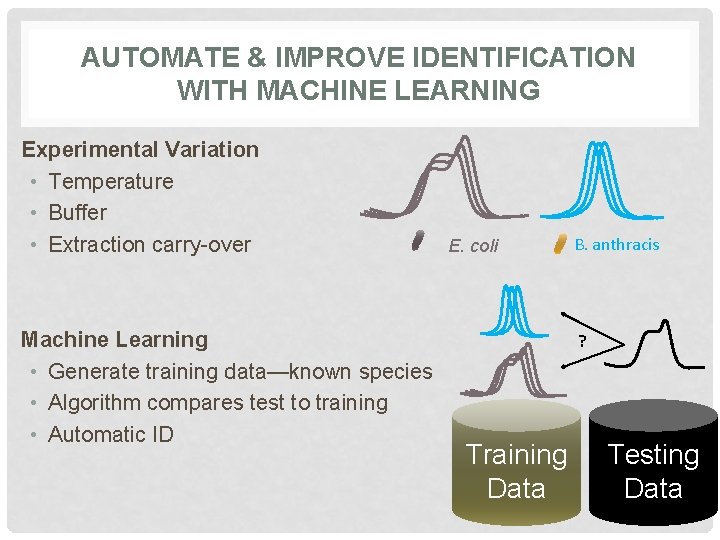
AUTOMATE & IMPROVE IDENTIFICATION WITH MACHINE LEARNING Experimental Variation • Temperature • Buffer • Extraction carry-over Machine Learning • Generate training data—known species • Algorithm compares test to training • Automatic ID E. coli B. anthracis ? Training Data Testing Data
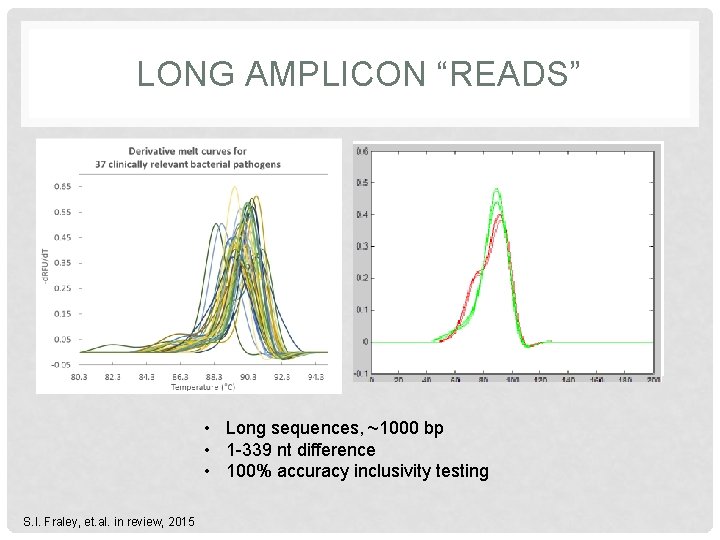
LONG AMPLICON “READS” • Long sequences, ~1000 bp • 1 -339 nt difference • 100% accuracy inclusivity testing S. I. Fraley, et. al. in review, 2015
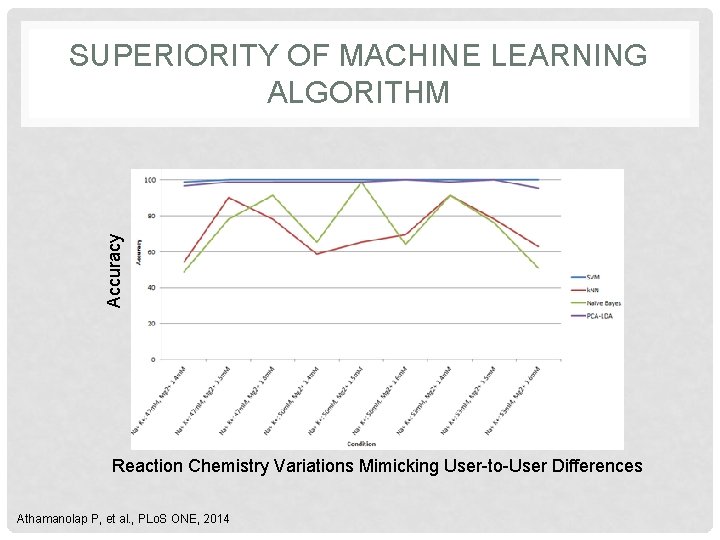
Accuracy SUPERIORITY OF MACHINE LEARNING ALGORITHM Reaction Chemistry Variations Mimicking User-to-User Differences Athamanolap P, et al. , PLo. S ONE, 2014
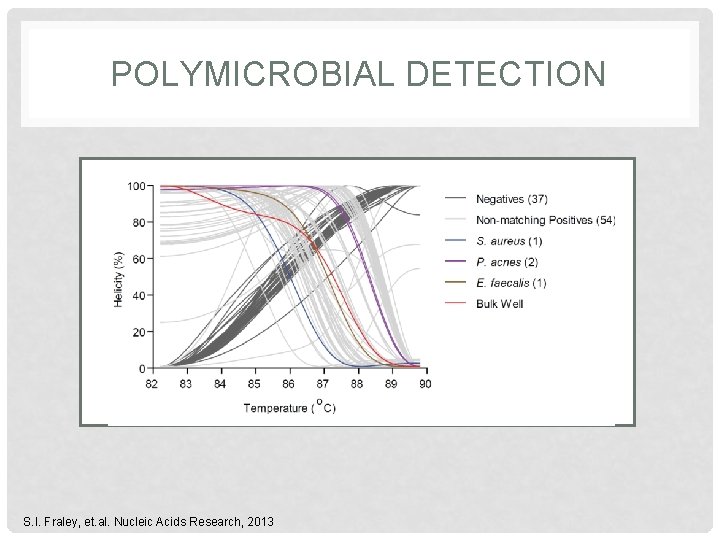
POLYMICROBIAL DETECTION S. I. Fraley, et. al. Nucleic Acids Research, 2013
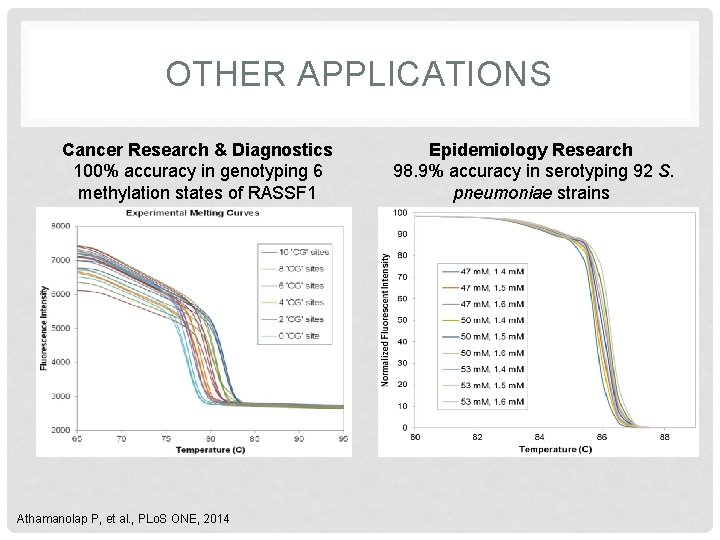
OTHER APPLICATIONS Cancer Research & Diagnostics 100% accuracy in genotyping 6 methylation states of RASSF 1 Athamanolap P, et al. , PLo. S ONE, 2014 Epidemiology Research 98. 9% accuracy in serotyping 92 S. pneumoniae strains
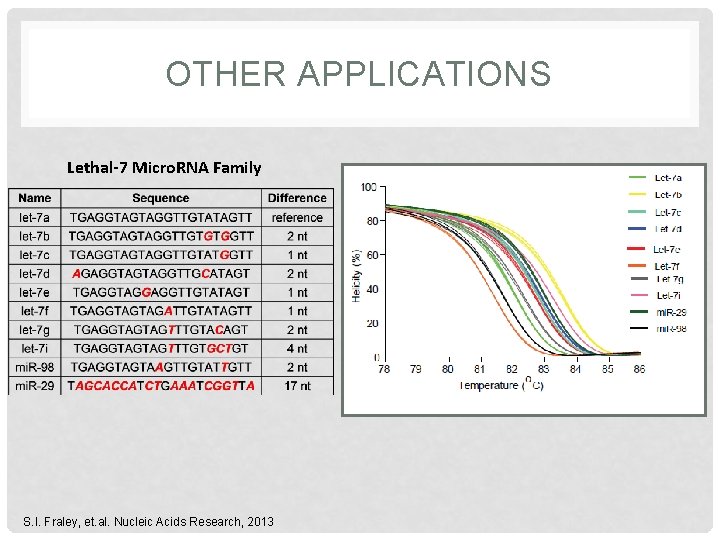
OTHER APPLICATIONS Lethal-7 Micro. RNA Family S. I. Fraley, et. al. Nucleic Acids Research, 2013
SUMMARY Automated, scalable, rapid, quantitative, inexpensive sequence identification • Universal PCR • Nucleic Acid Melting • Machine Learning • Microfluidics Tunable for diverse applications • Bacterial pathogens • Micro. RNA biomarkers • Cancer gene methylation signatures Future Goals • Portability • Integrating automated sample preparation • Expand database, 200 clinically relevant bacterial pathogens • Sequence melt prediction for training in silico and emerging pathogens

ACKNOWLEDGEMENTS v v v Samuel Yang, Emergency Medicine, Johns Hopkins, now at Stanford Jeff Wang, Biomedical Engineering, Johns Hopkins Richard Rothman, Emergency Medicine, Johns Hopkins Charlotte Gaydos, Emergency Medicine, Johns Hopkins Karen Carroll, Pathology, Johns Hopkins v Pornpat Apthamanolap, Johns Hopkins v Justin Hardick, Johns Hopkins v Billie Maseck, Johns Hopkins v Helena Zec, Johns Hopkins Fraley Lab v Daniel Ortiz-Velez, UC San Diego v Sinead Hawker, UC San. Diego Contact Information: Sifraley@ucsd. edu
- Slides: 18