En Tex GTEx comparison Anna Vlasova 09082017 ENTEx













- Slides: 17

En. Tex -GTEx comparison Anna Vlasova 09/08/2017

ENTEx 97 samples from 27 tissue subregions (SMTSD values) Reads mapped to the GRCh 38, gencode 24, STAR+RSEM pipeline GTEx 95 samples from 30 tissue subregions (SMTSD values) Reads mapped to the GRCh 38, gencode 26, STAR+RSEM+RNASe. QC pipeline GTEx only tissues: Whole Blood Artery – Tibial Esophagus – Muscularis Artery - Coronary ENTEx only tissues: Peyers patch In total there are 79 matched samples, both tissue and individual. For 7 samples En. Tex has replicates. Note: En. Tex and GTEx samples have different notation of the tissues, i. e. esophagus mucosa == Esophagus – Mucosa esophagus squamous epithelium =? Esophagus – Mucosa

Number of genes with TPM>=1 per sample Entex data – 86 samples, GTEx – 79 samples, matched tissues

Number of genes with TPM>=1, summary by study Entex data – 86 samples, GTEx – 79 samples, matched tissues

t-SNE clustering, using protein-coding genes, 21, 189 genes

t-SNE clustering, using tissue specific genes from GTEx, 2, 908 genes

Clustering of the samples based on the pearson correlation, protein coding genes

Pearson correlation for paired samples only, 72 samples, for protein coding genes Calculated on log 2(TPM+0. 1)

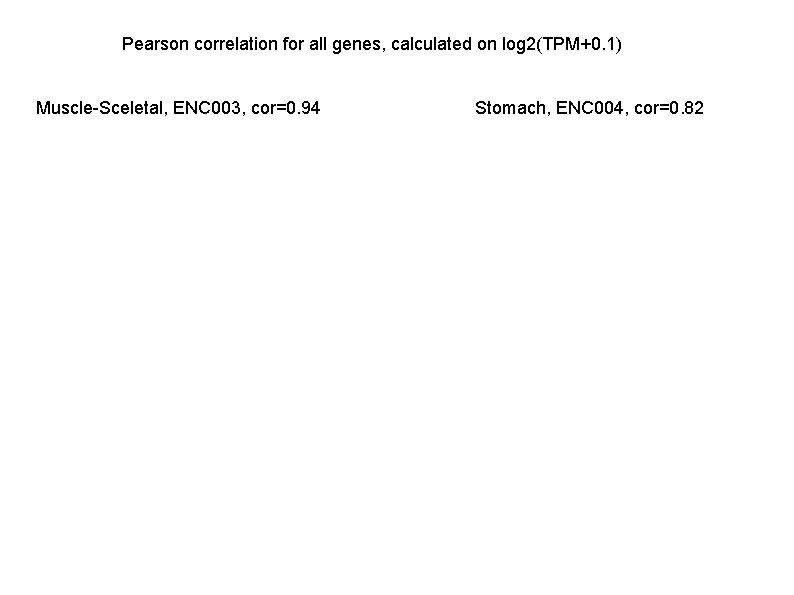
Pearson correlation for all genes, calculated on log 2(TPM+0. 1) Muscle-Sceletal, ENC 003, cor=0. 94 Stomach, ENC 004, cor=0. 82

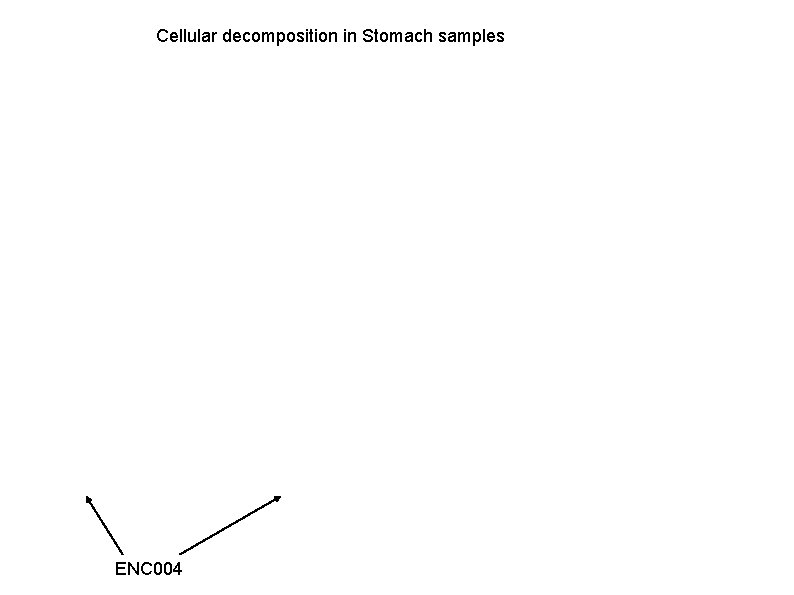
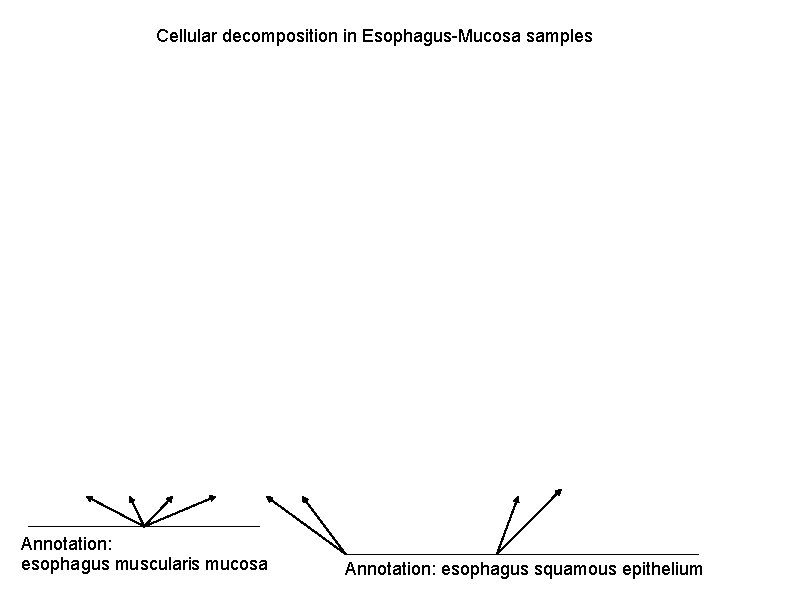
Cellular decomposition in En. Tex and GTEx samples, by applying Alessandra's method.

Cellular decomposition in Stomach samples ENC 004

Cellular decomposition in Esophagus-Mucosa samples Annotation: esophagus muscularis mucosa Annotation: esophagus squamous epithelium

Differential gene expression(DGE) between Gtex and Entex samples Lowly expressed genes Number of DGE genes between studies when using all tissues: Up-regulated in Entex – 1, 059 Up-regulated in GTEx – 1, 587 edge. R, log. FC>=2, FDR<=0. 05

70 genes differentially expressed between GTEx and En. TEx in all tissues. log 10(TPM+0. 1)

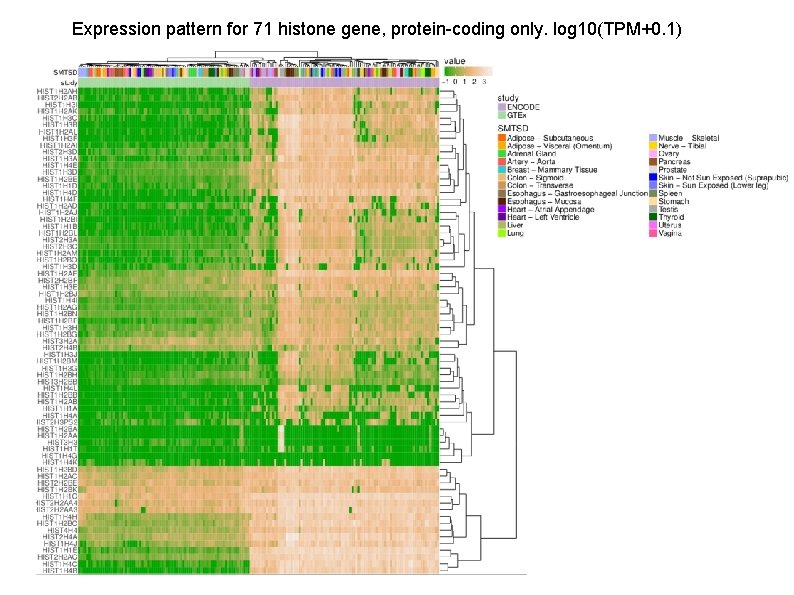
Genes expressed only in Entex samples those reported lack a poly. A tail: Histone genes (HIST 4 H 4, HIST 1 H 2 BC, HIST 2 H 2 AC, and some more), the RNA component of RNase P (RPPH), the RNA component of telomerase (TERC) Zinc finger protein ZNF 460 Genes expressed only in GTEx samples: Many antisense and TEC(To be Experimentally Confirmed) genes Protein coding genes with tag "overlapping_locus" Located in chr. M Yang et al. , Genome wide characterization of non-polyadenylated RNAs, Genome biology 2011

Expression pattern for 71 histone gene, protein-coding only. log 10(TPM+0. 1)

Tissue specific genes, 4, 749 genes, log 10(TPM+0. 1)