Empirical Validation of the Effectiveness of Chemical Descriptors








- Slides: 21

Empirical Validation of the Effectiveness of Chemical Descriptors in Data Mining Kirk Simmons Du. Pont Crop Protection Stine-Haskell Research Center 1090 Elkton Road Newark, DE 19711 kirk. a. simmons@usa. dupont. com

The Study • Purpose • Strategy – Methods – Metrics • Results • Practical Application • Conclusions

Purpose • Chemical Structure Conference (1996) – Holland – – Data mining/similarity methodologies reported Used numerous descriptor sets No standard datasets Comparisons difficult • Comparative study of chemical descriptors across varied biology

Strategy • Systematically evaluate descriptors within a compound dataset across multiple biological endpoints • All compounds have experimentally measured endpoints • Diversity of biological endpoints – In-Vitro (receptor affinity, enzyme inhibition) – In-Vivo (insect mortality) • Explored nine common descriptor sets • Train and then use model to forecast a validation set

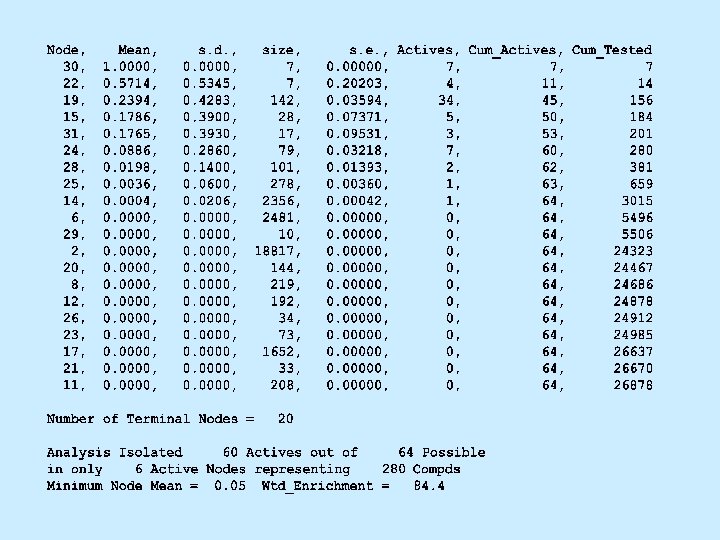
Methods • Four In-Vitro assays – 48 K compound dataset for training – Corporate database for validation • Two In-Vivo assays – 75 -100 K compound datasets – Randomly divided into training and validation subsets • Recursive Partitioning - analytic method – Appropriate method for HTS data – Selected statistically conservative inputs (p-tail < 0. 01)



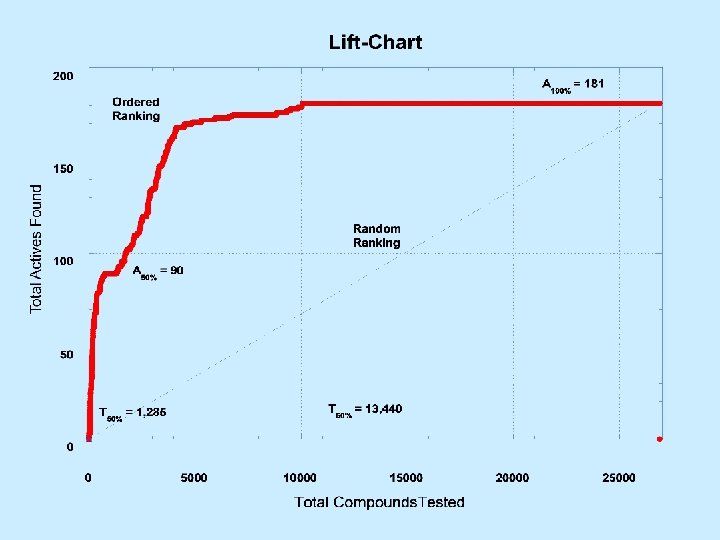
Metrics • 4 -way Interaction – Analytic Method, Compound Set, Biology, and Descriptors • Efficiency of analysis (Lift Chart) – Fraction of Actives found/Fraction of Dataset tested – Rewards efficiency only • Effectiveness of analysis (Composite Score) – Fraction of Actives found x Efficiency – Rewards efficiency as well as completeness




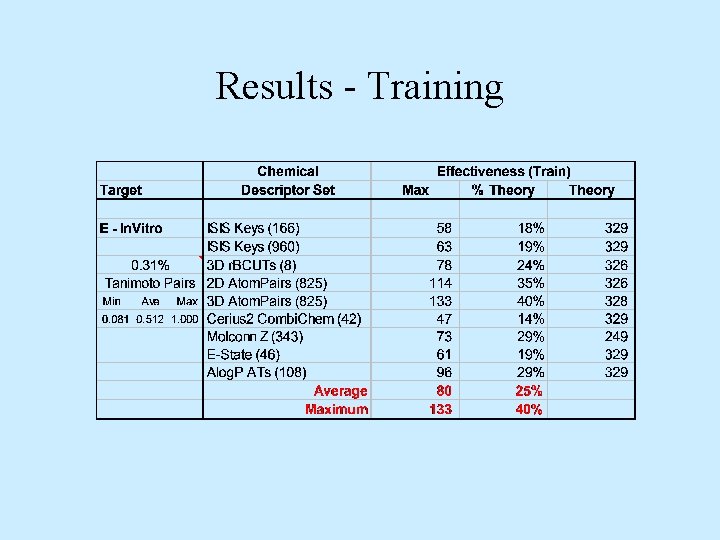
Results - Training

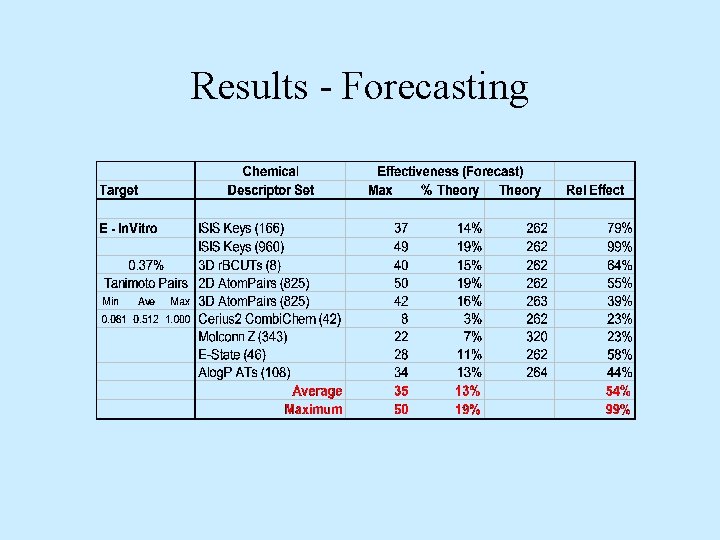
Results - Forecasting

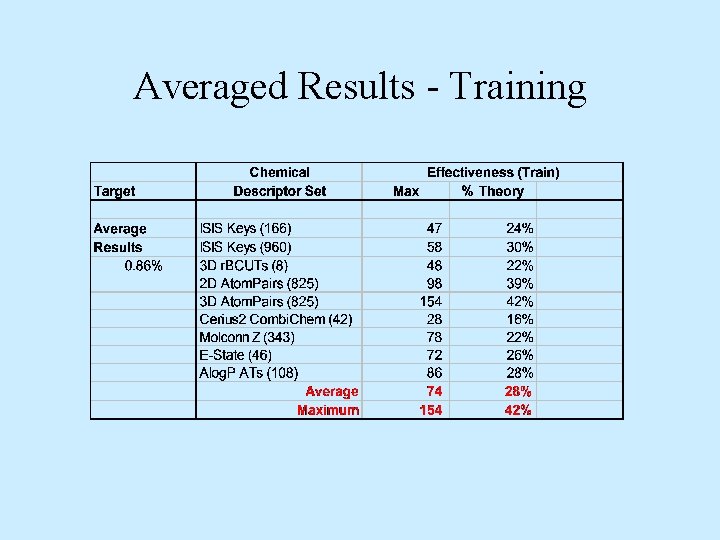
Averaged Results - Training

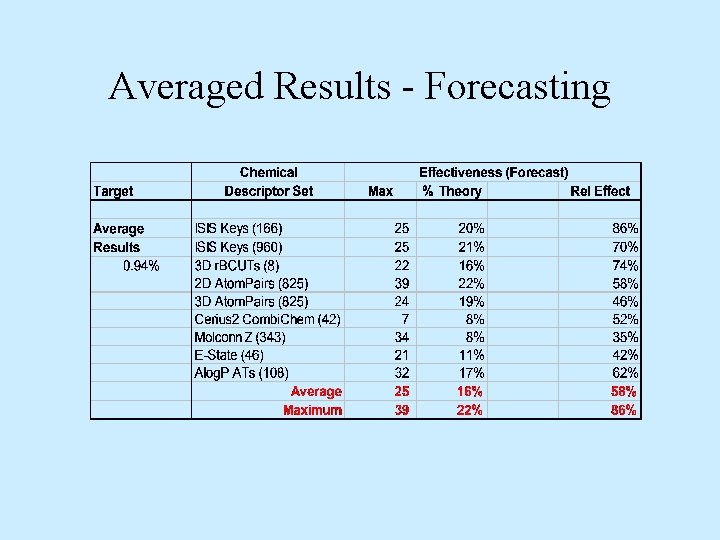
Averaged Results - Forecasting

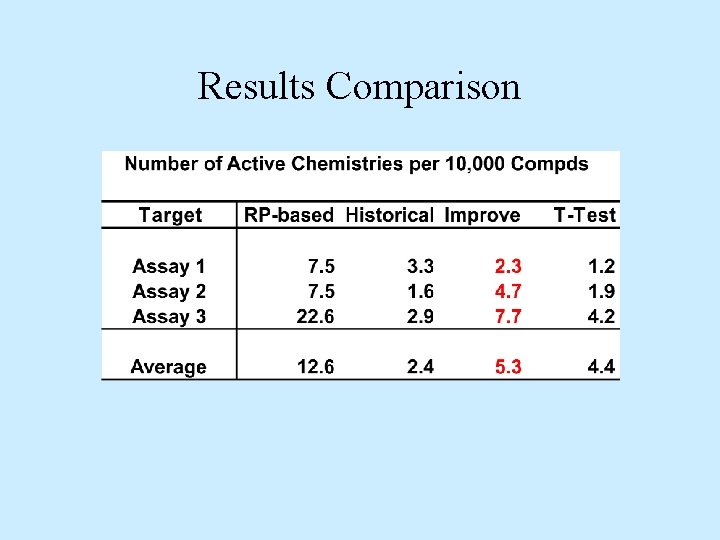
Practical Application • RP-based models using screening data on 3 targets – Activity treated as active/inactive – Diverse. Solutions. R BCUT descriptors • RP-models used to forecast vendor compounds (1 M) • Selected compounds purchased/screened – Hit-rates improved 530% over training sets – New structures and improved activity

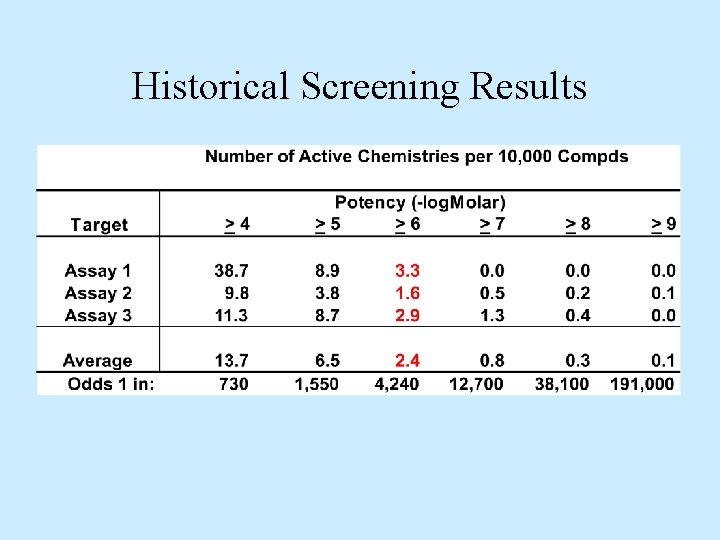
Historical Screening Results

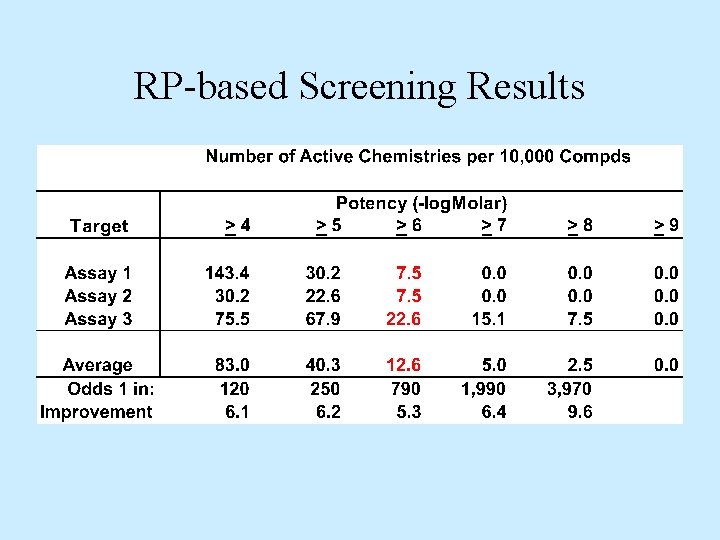
RP-based Screening Results

Results Comparison

Conclusions • Not all chemical descriptors equally effective – Whole molecule property-based less effective – Chemical feature-based appear more effective • Training models effectiveness – Averaged 28% of theory – Room for 4 -fold improvement • Validation models effectiveness – Averaged 16% of theory – Room for 6 -fold improvement

Acknowledgements • Dr. Linrong Yang, FMC Corporation – Completed the work • FMC Corporation – Release of the results • • Prof. Peter Willett, University of Sheffield Prof. Alex Tropsha, University of North Carolina Prof. Doug Hawkins, University Minnesota Du. Pont Corporation