ELECTRON CRYSTALLOGRAPHY Its role in proteomics Present and







- Slides: 35

ELECTRON CRYSTALLOGRAPHY: Its role in proteomics, Present and future Kenneth H. Downing Lawrence Berkeley National Laboratory

Resolution of present microscopes -- ~1Å, but much worse for biology Fundamental problem in obtaining biological data by EM is radiation damage Exposure ~ 10 electron/Å2, Noise ~ 30% in 1 -Å pixel Improve signal-to-noise ratio by averaging many equivalent images

Crystals provide a large number of equivalent images in a single shot -- all in same orientation, so easy to average Examples of structures solved by Electron crystallography: Results, limitations, prospects…

Tubulin: A cytoskeletal protein of eukaryotic cells that is essential for many functions

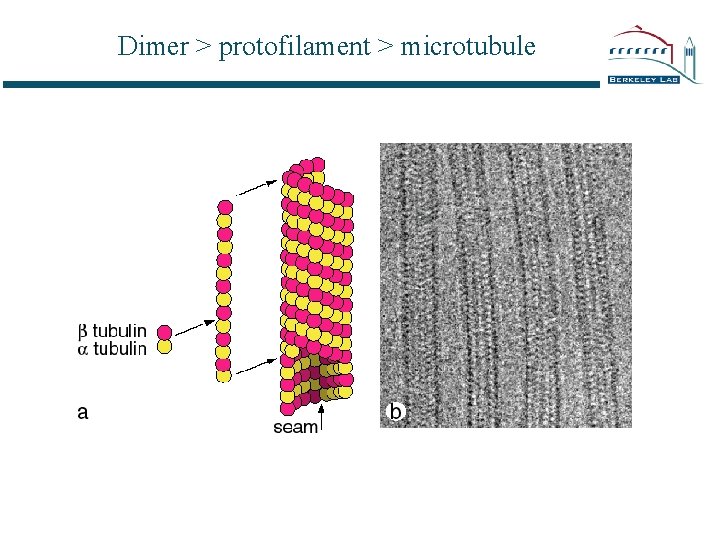
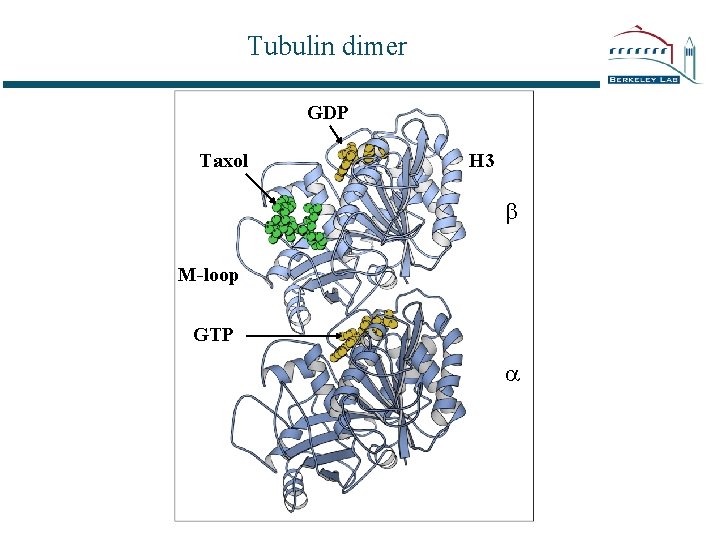
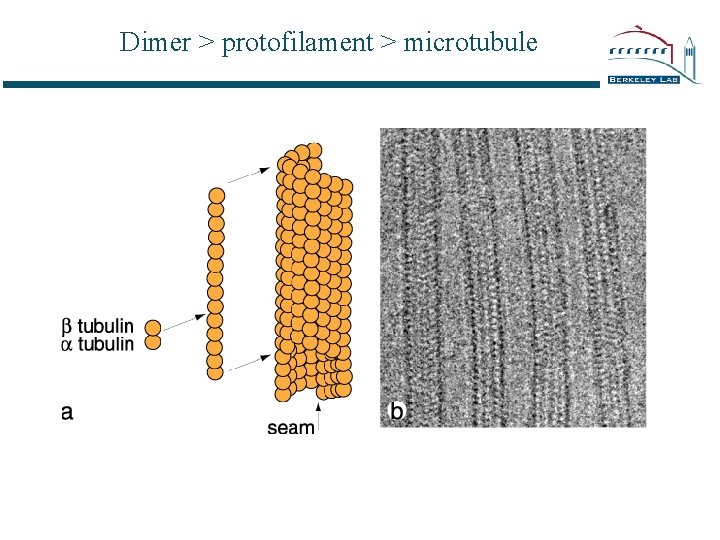
Dimer > protofilament > microtubule

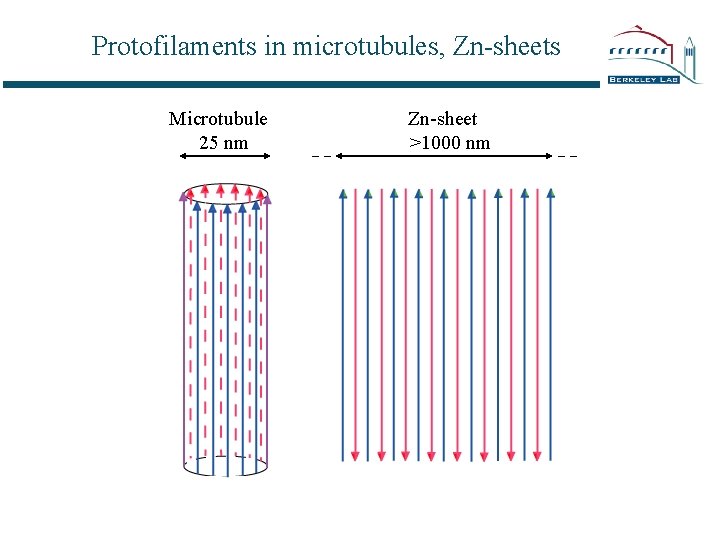
Protofilaments in microtubules, Zn-sheets Microtubule 25 nm Zn-sheet >1000 nm

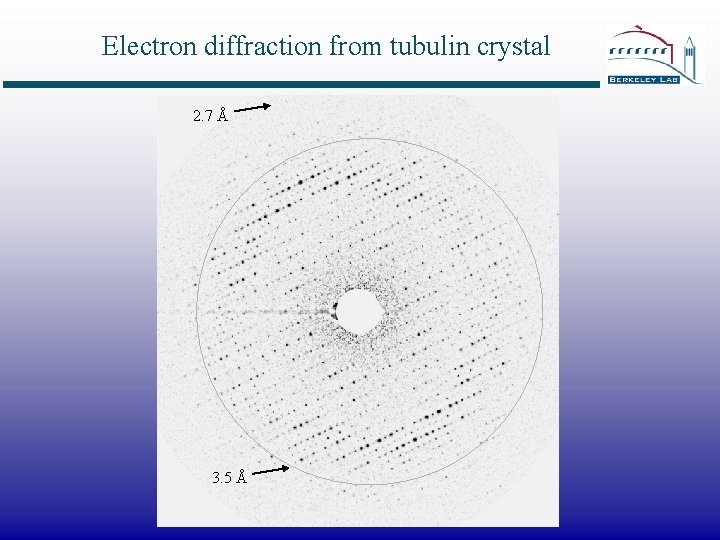
Electron diffraction from tubulin crystal 2. 7 Å 3. 5 Å

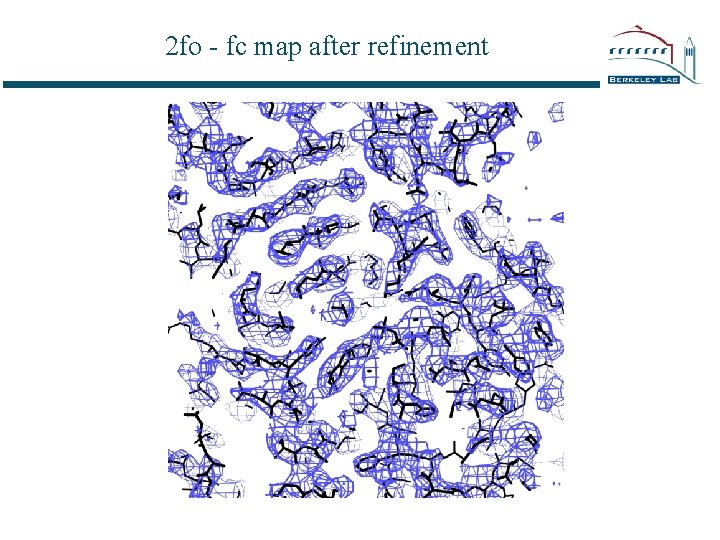
2 fo - fc map after refinement

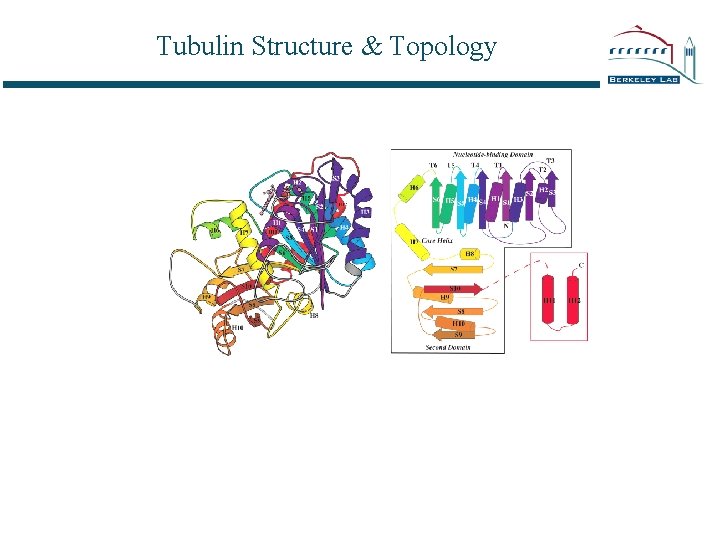
Tubulin Structure & Topology

Tubulin dimer GDP Taxol H 3 b M-loop GTP a

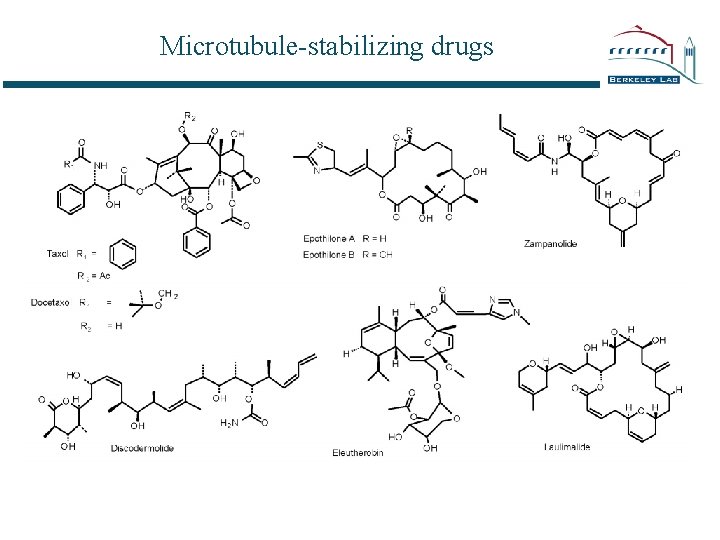
Tubulin - drug interactions Drugs that interfere with microtubule dynamics can stop cell division Taxol stabilizes microtubules -- as do several other drugs: epothilones sarcodictyin / eleutherobin discodermolide many Taxol (paclitaxel) analogues • These can be studied by diffraction methods

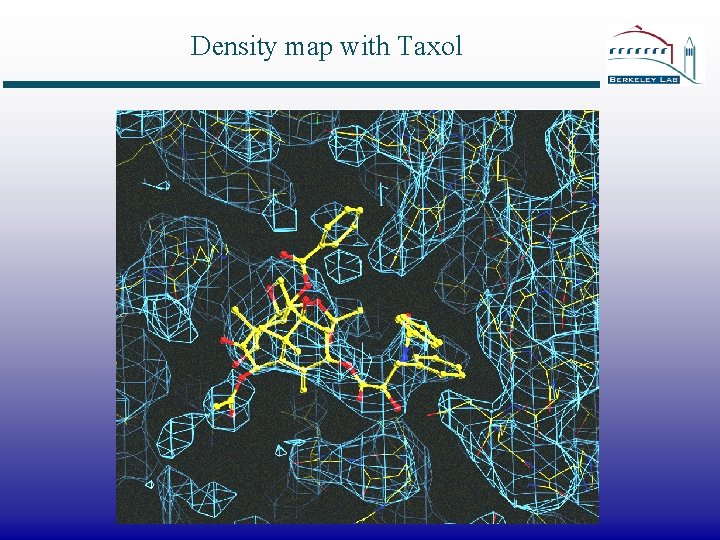
Density map with Taxol

Microtubule-stabilizing drugs

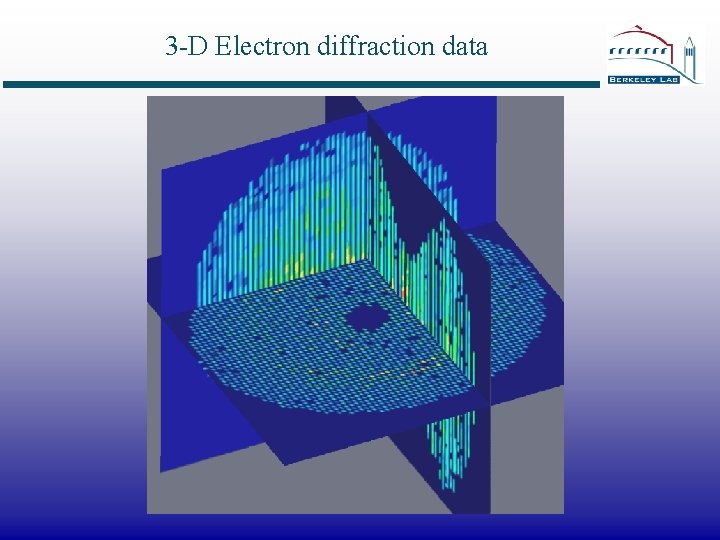
3 -D Electron diffraction data

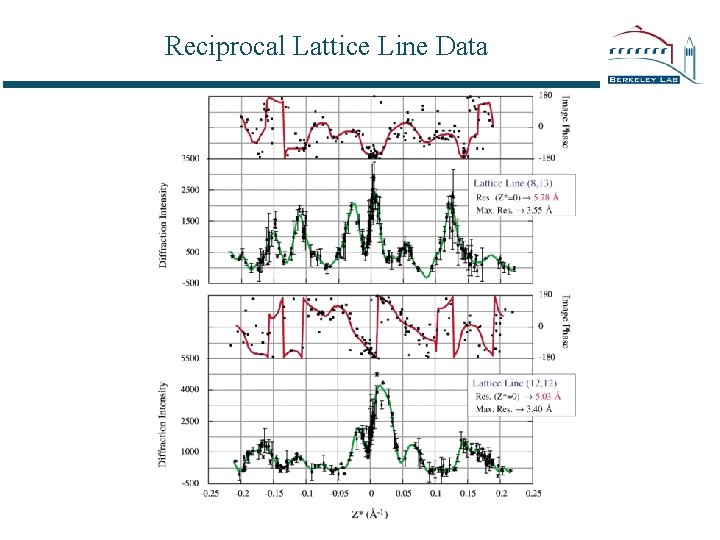
Reciprocal Lattice Line Data

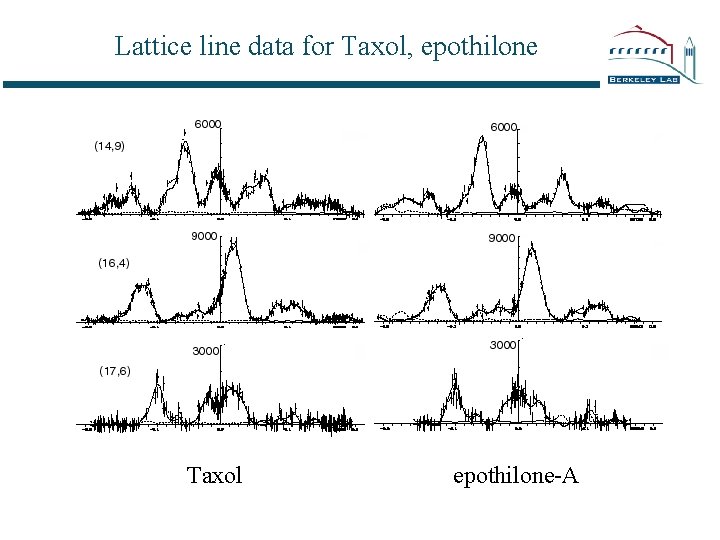
Lattice line data for Taxol, epothilone Taxol epothilone-A

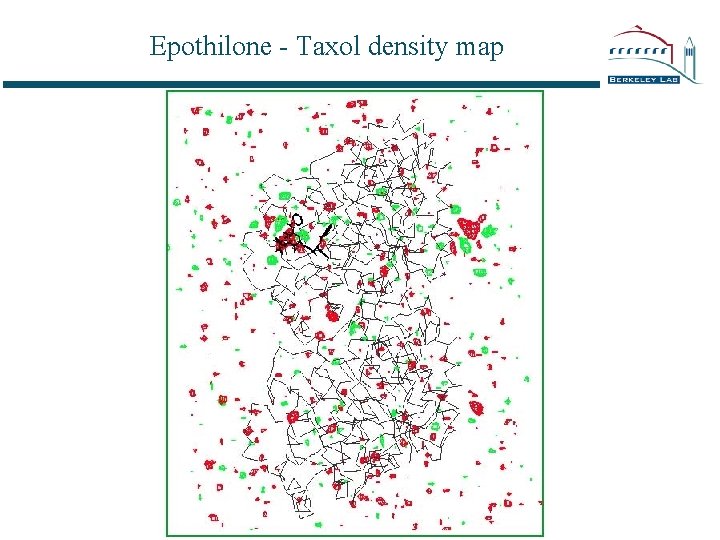
Epothilone - Taxol density map

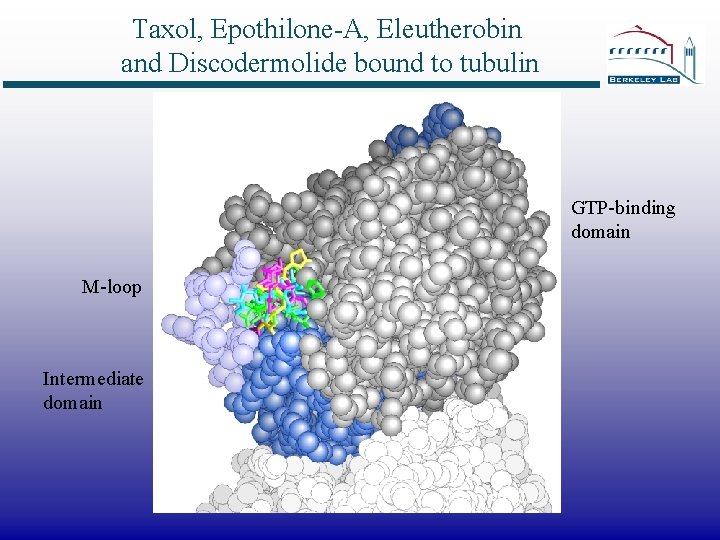
Taxol, Epothilone-A, Eleutherobin and Discodermolide bound to tubulin GTP-binding domain M-loop Intermediate domain

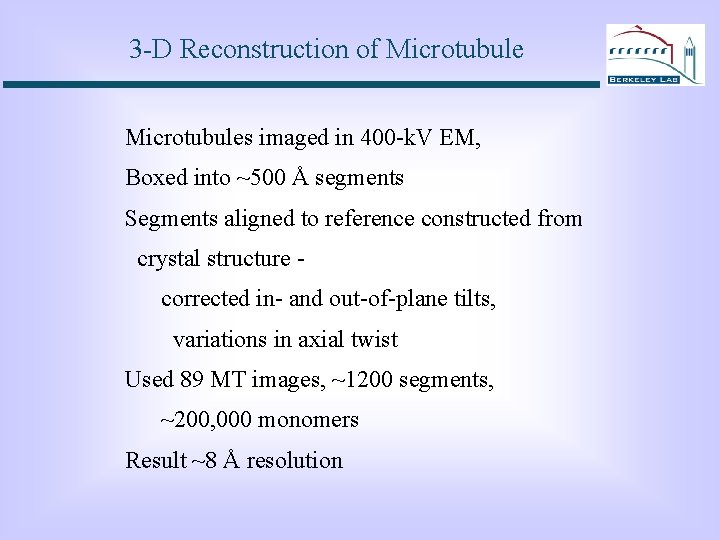
3 -D Reconstruction of Microtubules imaged in 400 -k. V EM, Boxed into ~500 Å segments Segments aligned to reference constructed from crystal structure corrected in- and out-of-plane tilts, variations in axial twist Used 89 MT images, ~1200 segments, ~200, 000 monomers Result ~8 Å resolution

Dimer > protofilament > microtubule

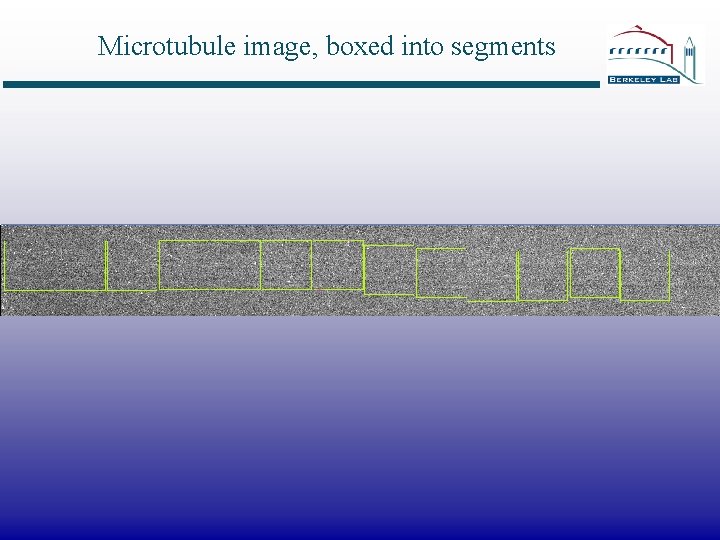
Microtubule image, boxed into segments

Microtubule map at 8 Angstroms

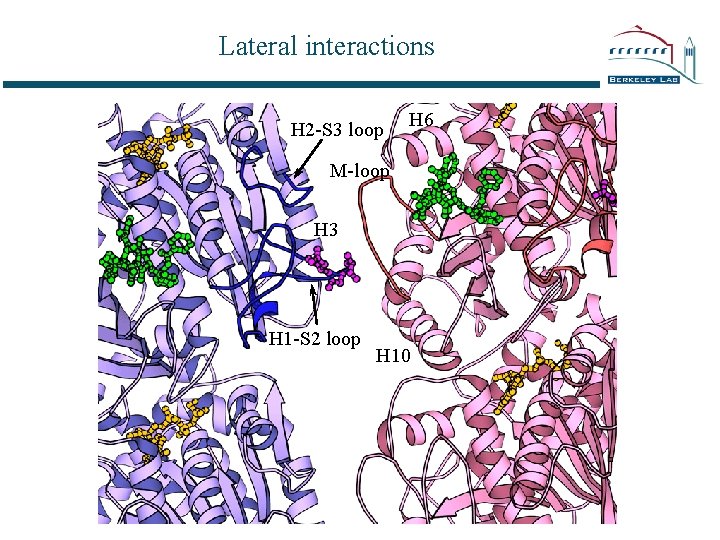
Lateral interactions H 2 -S 3 loop H 6 M-loop H 3 H 1 -S 2 loop H 10

Summary Tubulin structure solved by electron crystallography Drug interactions studied with diffraction data Microtubule structure by cryo-EM shows tubulin-tubulin interactions

BACTERIORHODOPSIN: A light-driven proton pump in bacteria Integral membrane protein Structural paradigm for all rhodopsins, G-protein coupled receptors

First 3 -D structure solved by electron crystallography (1990; resolution ~3. 5 Å) Refined structure, high resolution images ~1995 Higher-resolution 3 -D structures by EM, x-ray

BR in projection at 2. 6 Å resolution (Grigorieff, Beckmann, Zemlin 1995)

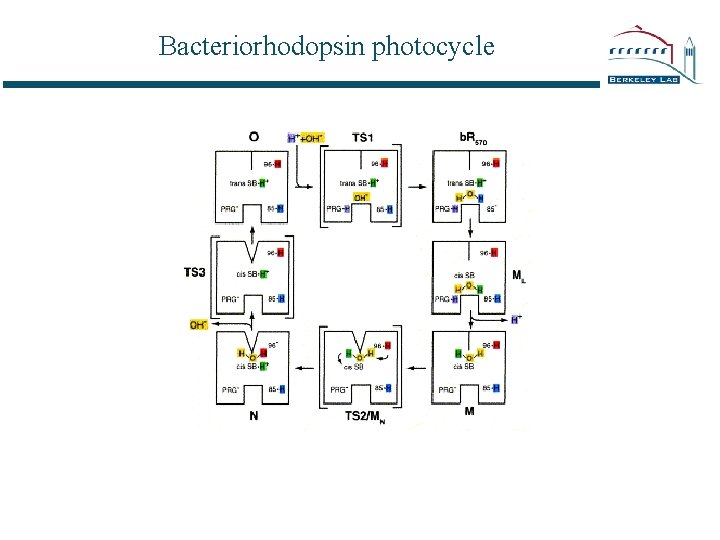
Bacteriorhodopsin photocycle

Summary Bacteriorhodopsin structure solved by electron crystallography Conformational changes studied by electron diffraction EM resolution extended to ~ 3 Å High resolution x-ray diffraction finally elucidated mechanism of proton pumping

How can EM compete with x-ray diffraction? • it shouldn’t compete! New instrumentation, along with continuing methods development -The keys to better and faster structure solutions Role for EM is mainly structures not amenable to x-ray

Our latest Electron Microscope

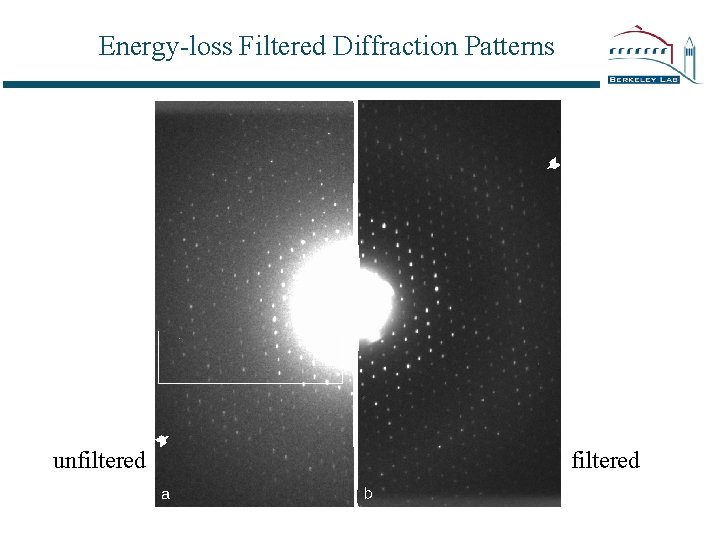
Energy-loss Filtered Diffraction Patterns unfiltered

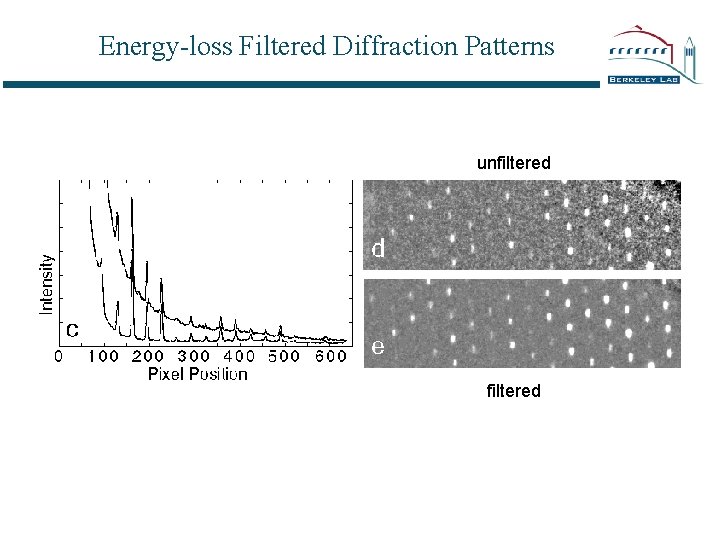
Energy-loss Filtered Diffraction Patterns unfiltered

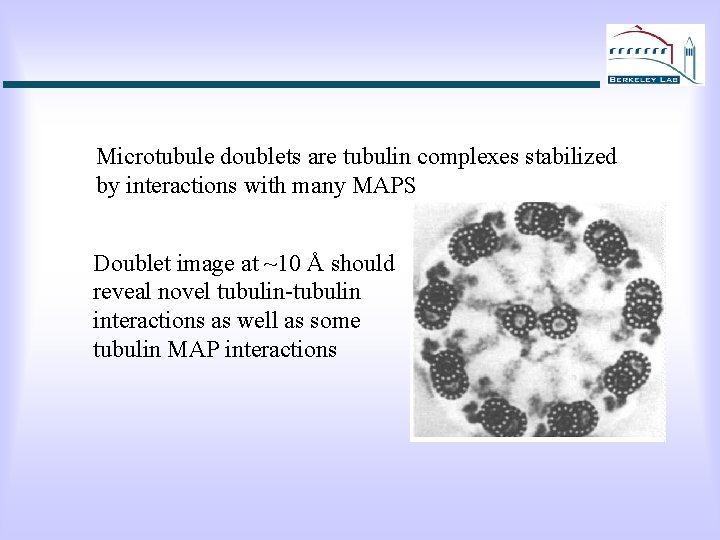
Microtubule doublets are tubulin complexes stabilized by interactions with many MAPS Doublet image at ~10 Å should reveal novel tubulin-tubulin interactions as well as some tubulin MAP interactions

The role of electron microscopy in proteomics: Electron crystallography gives single molecule structure at “atomic” resolution Ligand interactions and small conformational change can also be studied by crystallographic approaches EM is particularly good at studying large complexes