Efficient MapReducebased DBSCAN Algorithm with Optimized Data Partition
Efficient Map/Reduce-based DBSCAN Algorithm with Optimized Data Partition PRESENTED BY M Adithya 1 RN 09 IS 025 UNDER GUIDANCE OF Dr. M. V Sudhamani HOD, ISE RNSIT Department of Information Science and Engineering 2012 -2013 1
AGENDA § Introduction § DBSCAN Map/Reduce § Distributed Density-Based Clustering with Map. Reduce § Performance Evaluation § Conclusion Department of Information Science and Engineering 2012 -2013 2
ABSTRACT • DBSCAN density based clustering deals with noisy datasets • A Map/Reduce based DBSCAN to solve scalability problem (DBSCAN-MR) • Input data partitioned into smaller parts and parallel processed on hadoop. • PRBP to select partition boundaries Department of Information Science and Engineering 2012 -2013 3
INTRODUCTION • Discovering relationships and groups for decision making • Clustering techniques partition data points into groups • Traditional algorithms running on single processor face scalability problem • DBSCAN has ability of discovering structures with arbitrary shapes. Department of Information Science and Engineering 2012 -2013 4

Introduction contd. . • TI-DBSCAN uses triangle inequality • GRIDBSCAN constructs a grid that allocates data points into partitions. • DBSCAN-MR to address scalability problem. • Previous works have a global spatial index. • DBSCAN-MR which uses a distributed index along with PRBP optimizes load balance and execution efficiency. Department of Information Science and Engineering 2012 -2013 5
DBSCAN AND MAP/REDUCE • Given Radius ε(Eps), each object of a cluster has to contain at least a minimum number (Min. Pts) of neighborhoods. • If the Eps-neighborhood of p, NEps(p) has more points than Min. Pts, a new cluster C containing the points in NEps(p) is created. • Map: (k 1, v 1) →(k 2, v 2) Reduce: (k 2, v 2) →(k 2, v 3) Department of Information Science and Engineering 2012 -2013 6
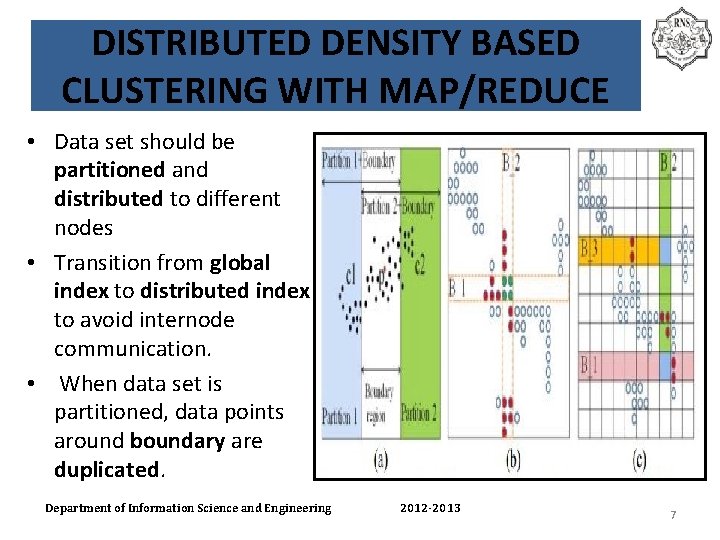
DISTRIBUTED DENSITY BASED CLUSTERING WITH MAP/REDUCE • Data set should be partitioned and distributed to different nodes • Transition from global index to distributed index to avoid internode communication. • When data set is partitioned, data points around boundary are duplicated. Department of Information Science and Engineering 2012 -2013 7
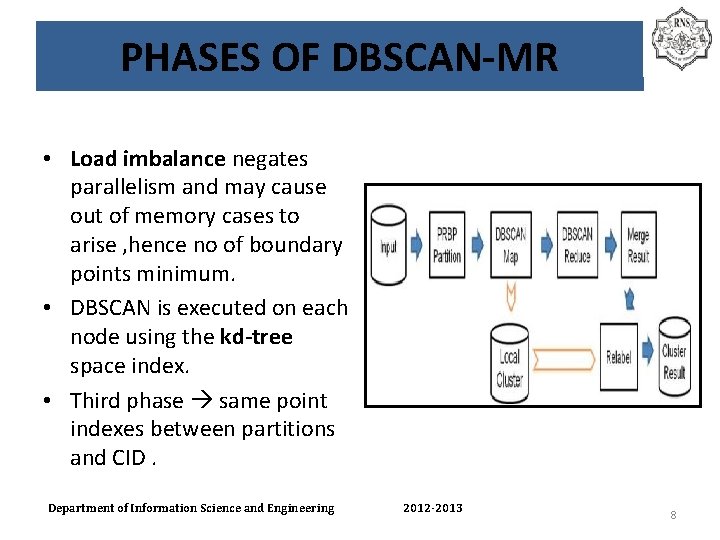
PHASES OF DBSCAN-MR • Load imbalance negates parallelism and may cause out of memory cases to arise , hence no of boundary points minimum. • DBSCAN is executed on each node using the kd-tree space index. • Third phase same point indexes between partitions and CID. Department of Information Science and Engineering 2012 -2013 8
PARTITION WITH REDUCED BOUNDARY POINTS • θ and β -> prevent unbalanced partitions and to avoid OOM condition. • Step 1 : Initializing slices for each dimension. • Step 2 : Calculating accumulative points for each slice. • Step 3 : Selecting the best slice to partition. • • • • • • Department of Information Science and Engineering Algorithm Partition with reduced boundary points (D, Eps, β, θ) Input: D: dataset; {Step I: initializing slices for each dimension} 1. p=build. Slice. Use 2 Eps(D, Eps, β, θ); 2. P. add (p); // P is a set of partitions {Step II: calculating accumulative points for each successive slice} 3. For each dimension d in p do 4. For each slice s in d do 5. Calculates the number of points s. count and the accumulative number of points s. total 6. End for {Step III: selecting the best slice to partition data} 7. For each partition p in P do 8. If p is not processed then 9. If partition. Use. Best. Slice (p, β, θ) is true // if return value is true, p is //split to two part in partition. Use. Best. Slice 10. Delete p from P; 11. End if 12. End if 13. End for 14. Return P 2012 -2013 9
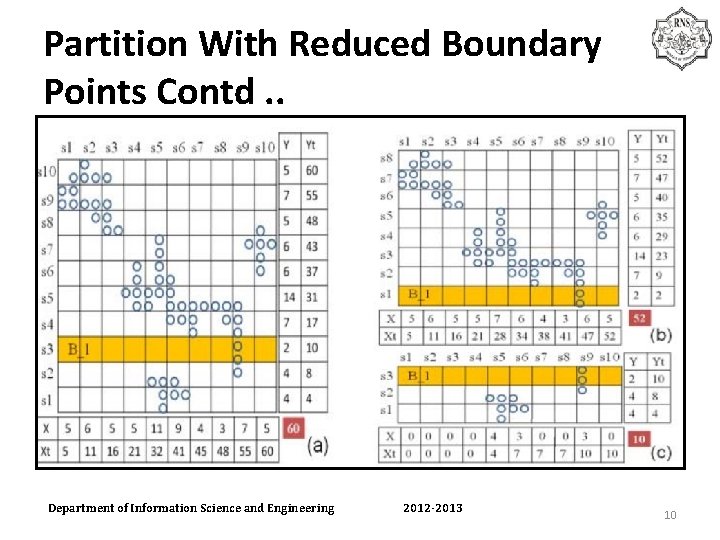
Partition With Reduced Boundary Points Contd. . Department of Information Science and Engineering 2012 -2013 10
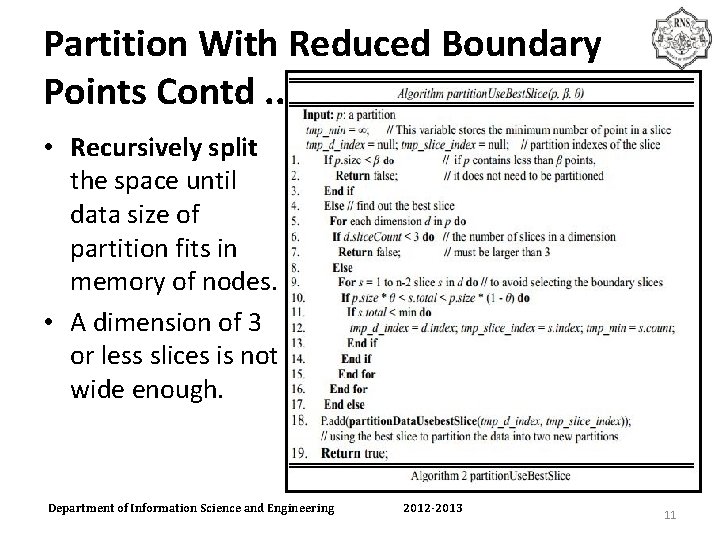
Partition With Reduced Boundary Points Contd. . • Recursively split the space until data size of partition fits in memory of nodes. • A dimension of 3 or less slices is not wide enough. Department of Information Science and Engineering 2012 -2013 11
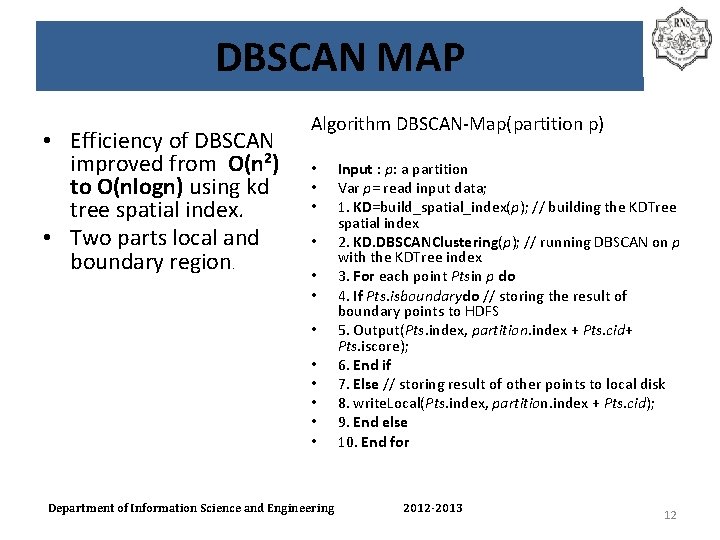
DBSCAN MAP • Efficiency of DBSCAN improved from O(n 2) to O(nlogn) using kd tree spatial index. • Two parts local and boundary region. Algorithm DBSCAN-Map(partition p) • • • Department of Information Science and Engineering Input : p: a partition Var p= read input data; 1. KD=build_spatial_index(p); // building the KDTree spatial index 2. KD. DBSCANClustering(p); // running DBSCAN on p with the KDTree index 3. For each point Ptsin p do 4. If Pts. isboundarydo // storing the result of boundary points to HDFS 5. Output(Pts. index, partition. index + Pts. cid+ Pts. iscore); 6. End if 7. Else // storing result of other points to local disk 8. write. Local(Pts. index, partition. index + Pts. cid); 9. End else 10. End for 2012 -2013 12
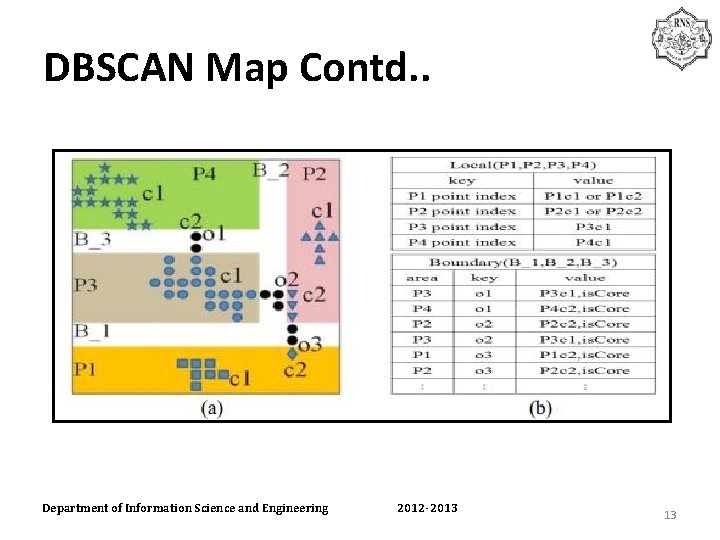
DBSCAN Map Contd. . Department of Information Science and Engineering 2012 -2013 13
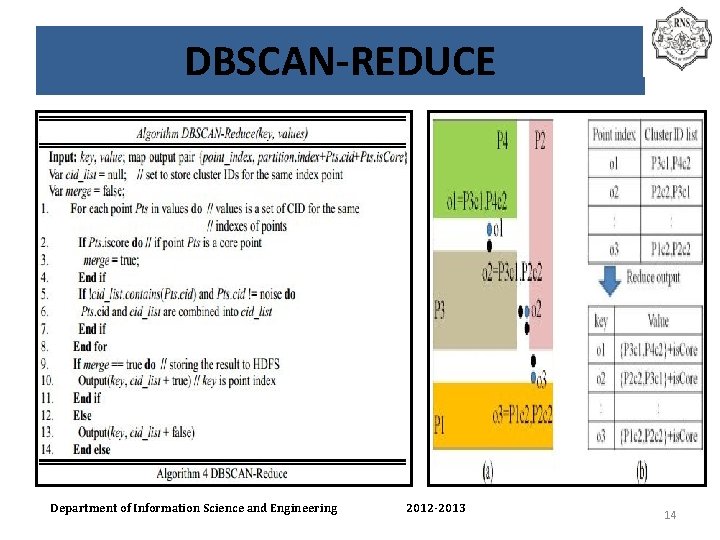
DBSCAN-REDUCE Department of Information Science and Engineering 2012 -2013 14
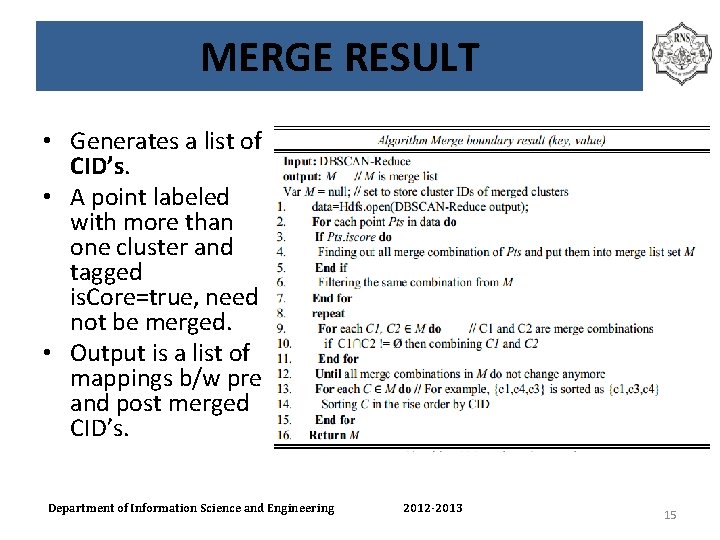
MERGE RESULT • Generates a list of CID’s. • A point labeled with more than one cluster and tagged is. Core=true, need not be merged. • Output is a list of mappings b/w pre and post merged CID’s. Department of Information Science and Engineering 2012 -2013 15
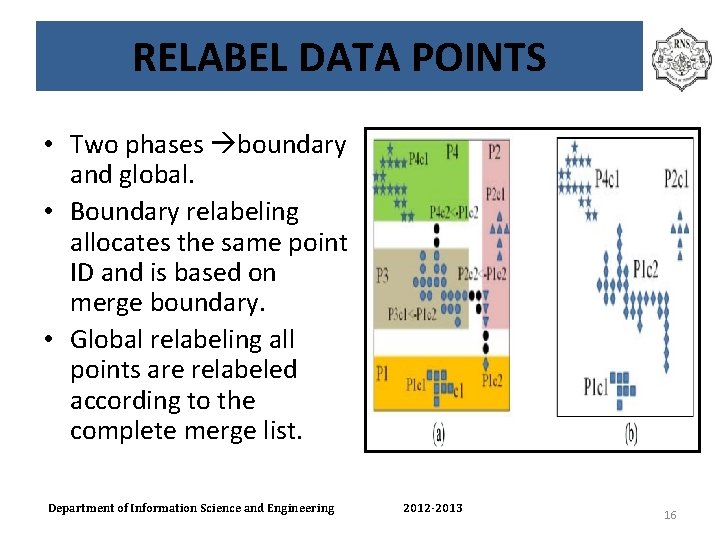
RELABEL DATA POINTS • Two phases boundary and global. • Boundary relabeling allocates the same point ID and is based on merge boundary. • Global relabeling all points are relabeled according to the complete merge list. Department of Information Science and Engineering 2012 -2013 16
PERFORMANCE EVALUATION • Algorithms compared are GRIDBSCAN, DBSCAN-MR-N and DBSCAN-MR • DBSCAN-MR-N uses only build. Slice. Use 2 eps method to select split region. • The hadoop cluster consists of 10 nodes and each node contains 4 Intel Xeon CPU 3. 00 GHz and 4 GB Ram running on Linux. Department of Information Science and Engineering 2012 -2013 17
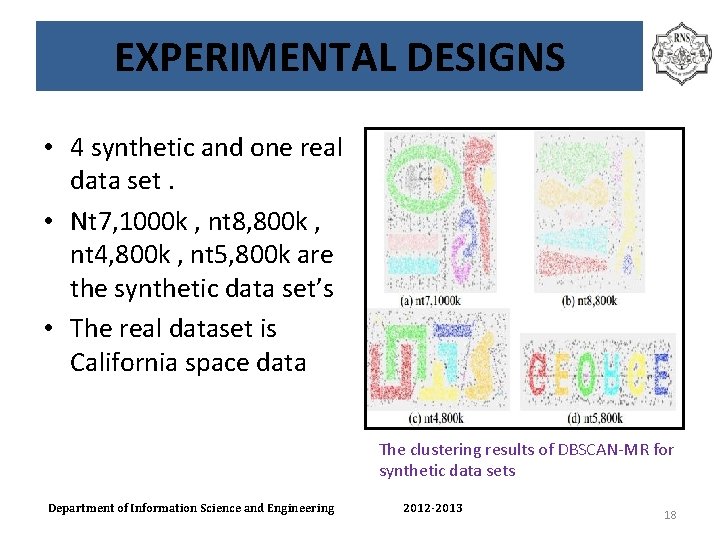
EXPERIMENTAL DESIGNS • 4 synthetic and one real data set. • Nt 7, 1000 k , nt 8, 800 k , nt 4, 800 k , nt 5, 800 k are the synthetic data set’s • The real dataset is California space data The clustering results of DBSCAN-MR for synthetic data sets Department of Information Science and Engineering 2012 -2013 18
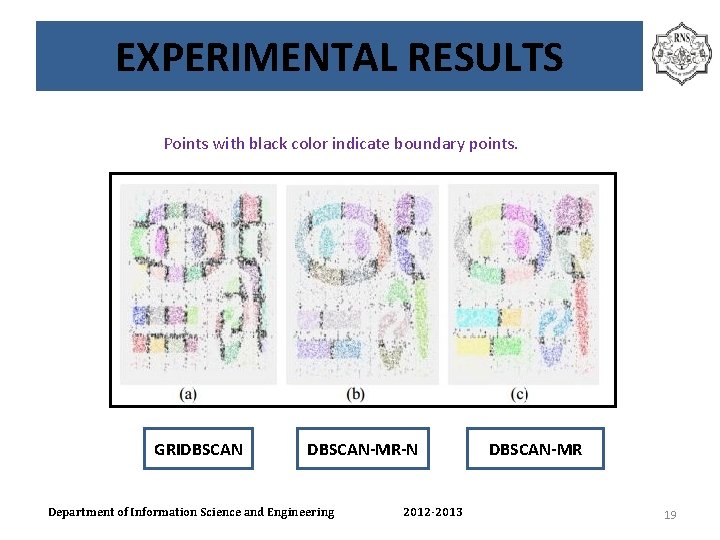
EXPERIMENTAL RESULTS Points with black color indicate boundary points. GRIDBSCAN-MR-N Department of Information Science and Engineering 2012 -2013 DBSCAN-MR 19
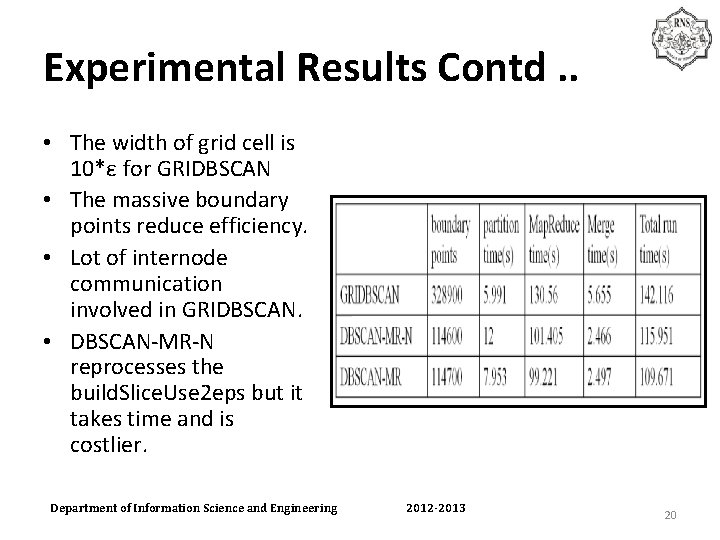
Experimental Results Contd. . • The width of grid cell is 10*ε for GRIDBSCAN • The massive boundary points reduce efficiency. • Lot of internode communication involved in GRIDBSCAN. • DBSCAN-MR-N reprocesses the build. Slice. Use 2 eps but it takes time and is costlier. Department of Information Science and Engineering 2012 -2013 20
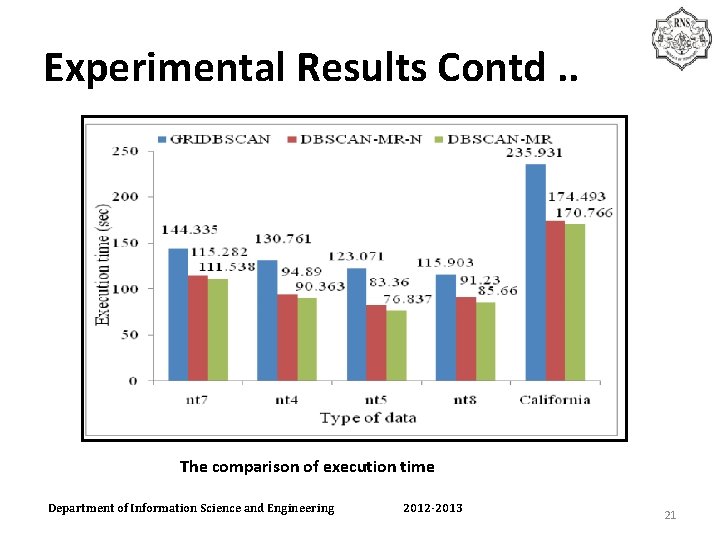
Experimental Results Contd. . The comparison of execution time Department of Information Science and Engineering 2012 -2013 21
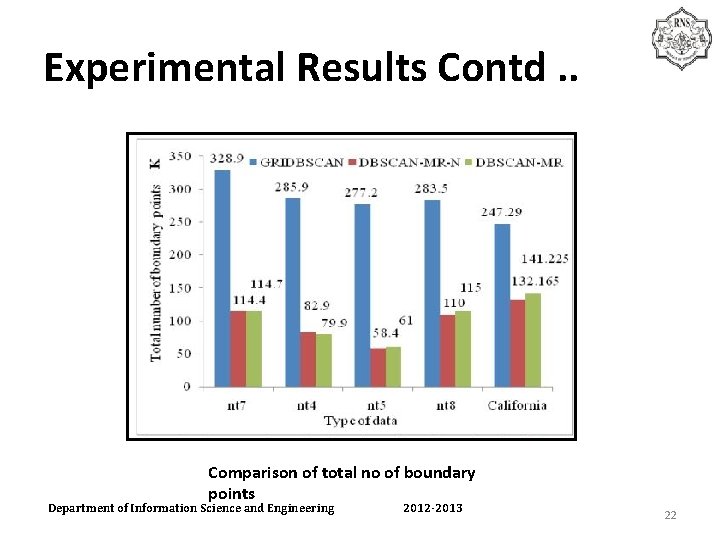
Experimental Results Contd. . Comparison of total no of boundary points Department of Information Science and Engineering 2012 -2013 22
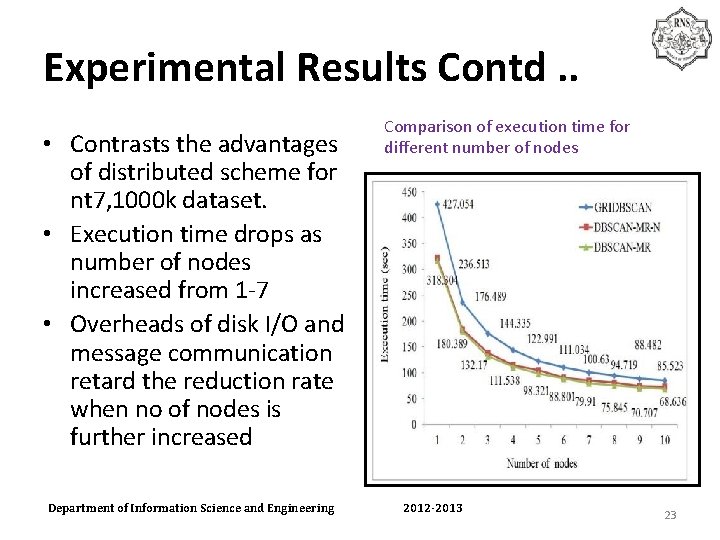
Experimental Results Contd. . • Contrasts the advantages of distributed scheme for nt 7, 1000 k dataset. • Execution time drops as number of nodes increased from 1 -7 • Overheads of disk I/O and message communication retard the reduction rate when no of nodes is further increased Department of Information Science and Engineering Comparison of execution time for different number of nodes 2012 -2013 23
CONCLUSION • DBSCAN-MR Increased the performance of DBSCAN by cloud computing technology. • PRBP balanced the load of each node and improved efficiency of entire framework. • Experimental results verified the high efficiency of DBSCAN-MR. Department of Information Science and Engineering 2012 -2013 24

Department of Information Science and Engineering 2012 -2013 25
- Slides: 25