Efficient discovery of diseasecausing genes using whole exome










- Slides: 39

Efficient discovery of disease-causing genes using whole exome sequencing Murim Choi, Ph. D Department of Biomedical Sciences College of Medicine Seoul National University

Overview 1. Introduction 2. Whole exome sequencing 3. Detecting somatic mutations - Benign tumor - Aldosterone-producing adenoma - Malignant tumor - Uterine serous carcinoma, TCGA - Mosaicism - Sturge-Weber syndrome

Human Genetics

The big questions of Human Genetics • How to detect genomic variants? • How to evaluate their functionality, esp. disease causality?

Eras of Disease Gene Discovery, as of 2013 1. Mendelian diseases by linkage analysis 2. Common diseases by common variants (SNPs) - GWAS 3. Mendelian diseases by rare variants - high-throughput sequencing 4. Common diseases by rare variants - high-throughput sequencing

Whole Exome Sequencing (WES) • Exome - Exons and exon-intron boundary sequences + mi. RNAs - ~1% of human genome (~35 Mb) - Responsible for majority of disease-traits • Whole Exome Sequencing - Cheaper and efficient - Requires less computing power/storage

One Solution - Whole Exome Sequencing (WES) Ng et al. , 2009 Nature Choi et al. , 2009 PNAS

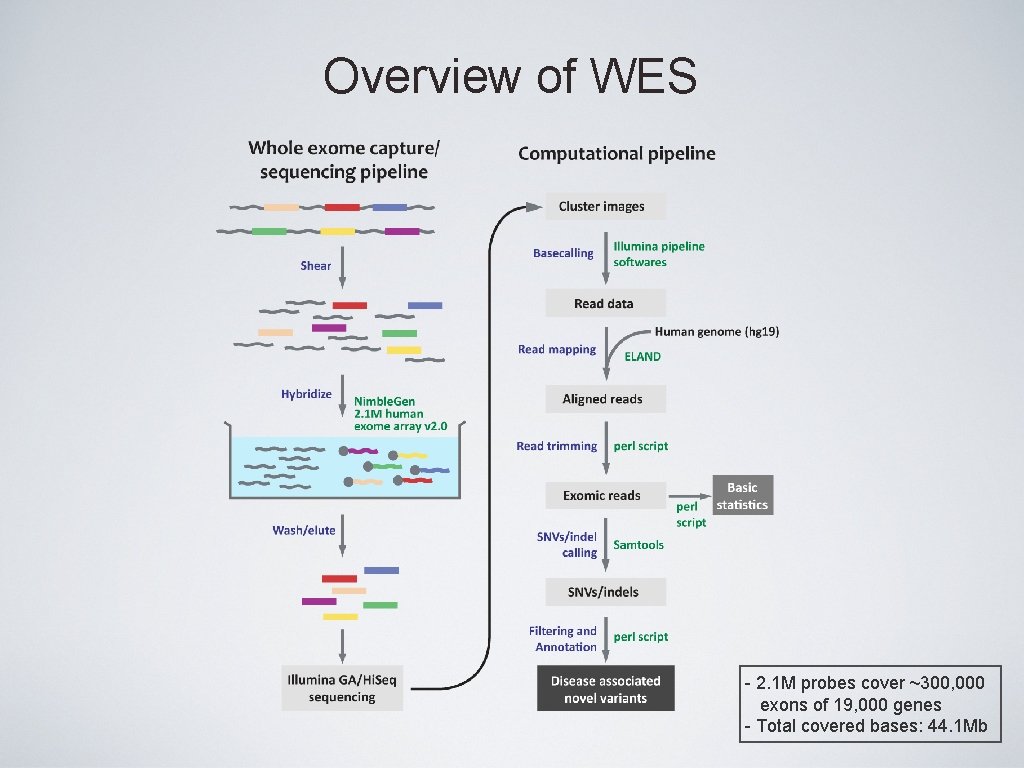
Overview of WES - 2. 1 M probes cover ~300, 000 exons of 19, 000 genes - Total covered bases: 44. 1 Mb

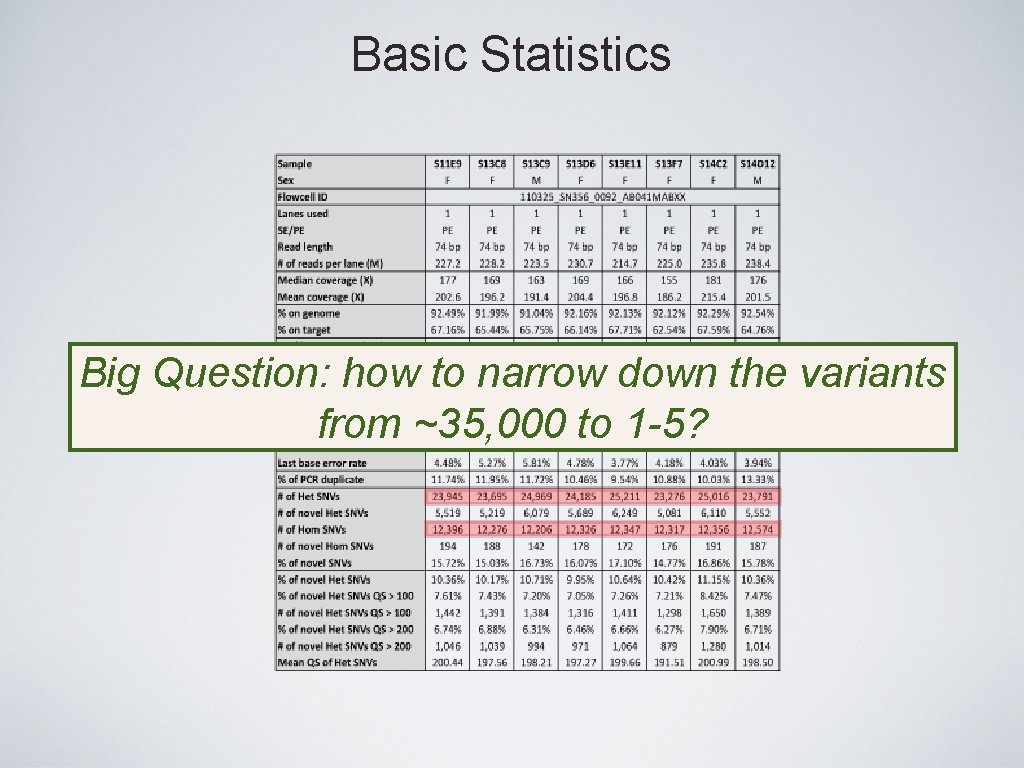
Basic Statistics Big Question: how to narrow down the variants from ~35, 000 to 1 -5?

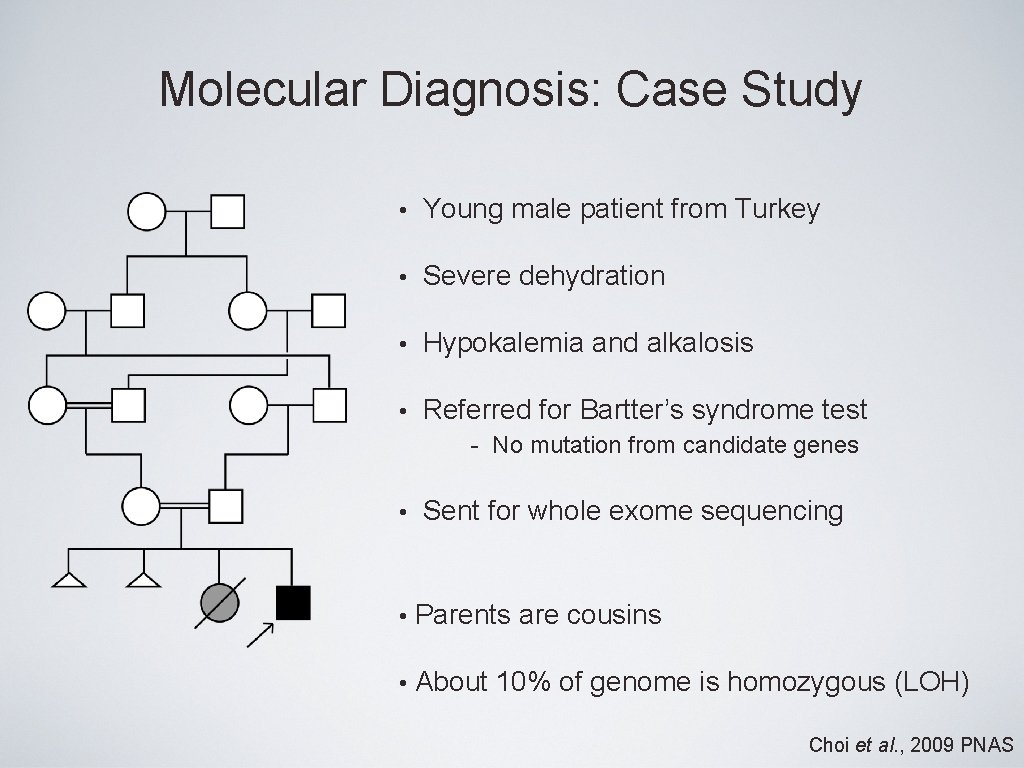
Molecular Diagnosis: Case Study • Young male patient from Turkey • Severe dehydration • Hypokalemia and alkalosis • Referred for Bartter’s syndrome test - No mutation from candidate genes • Sent for whole exome sequencing • Parents are cousins • About 10% of genome is homozygous (LOH) Choi et al. , 2009 PNAS

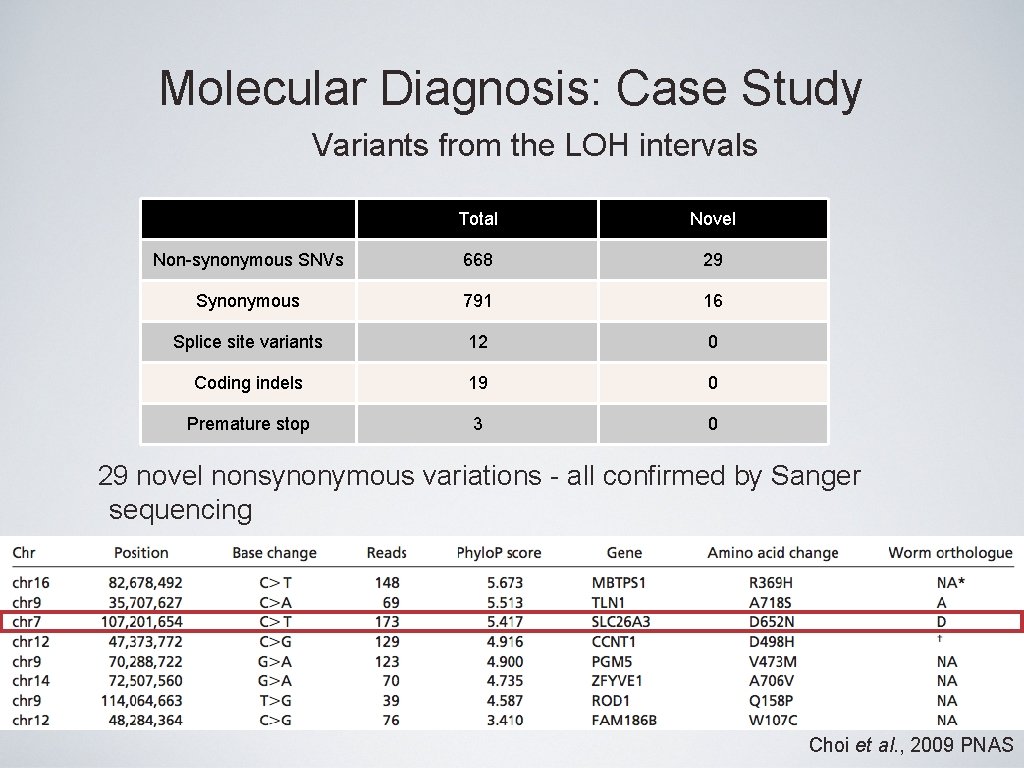
Molecular Diagnosis: Case Study Variants from the LOH intervals Total Novel Non-synonymous SNVs 668 29 Synonymous 791 16 Splice site variants 12 0 Coding indels 19 0 Premature stop 3 0 29 novel nonsynonymous variations - all confirmed by Sanger sequencing Choi et al. , 2009 PNAS

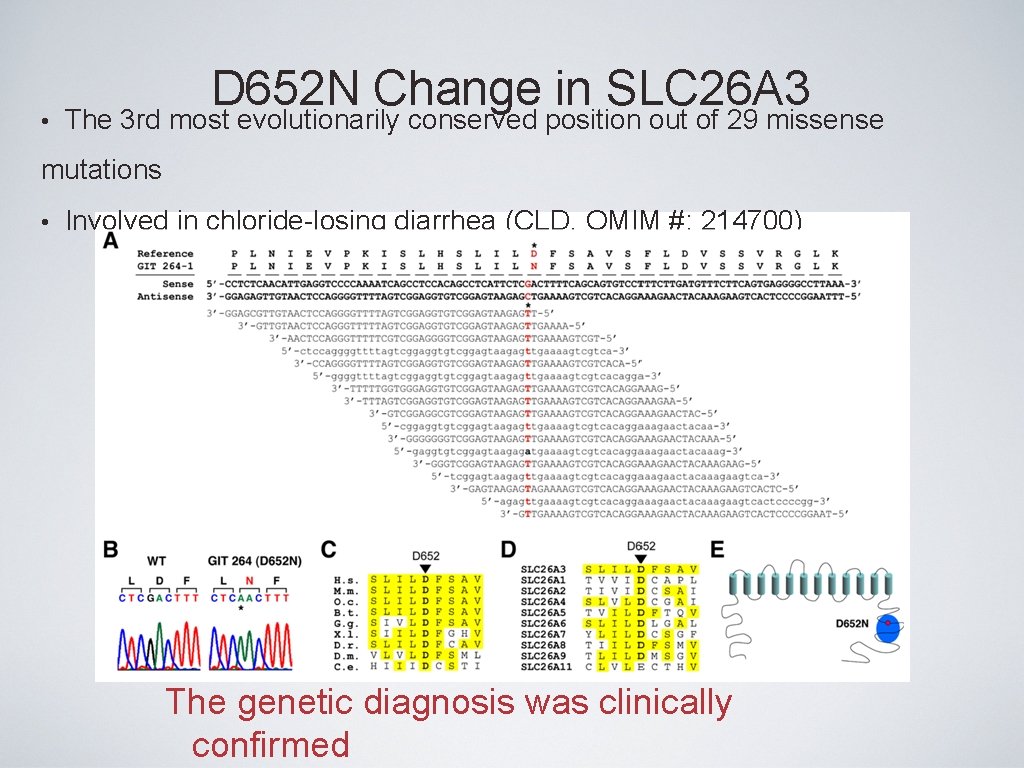
• D 652 N Change in SLC 26 A 3 The 3 rd most evolutionarily conserved position out of 29 missense mutations • Involved in chloride-losing diarrhea (CLD, OMIM #: 214700) The genetic diagnosis was clinically confirmed

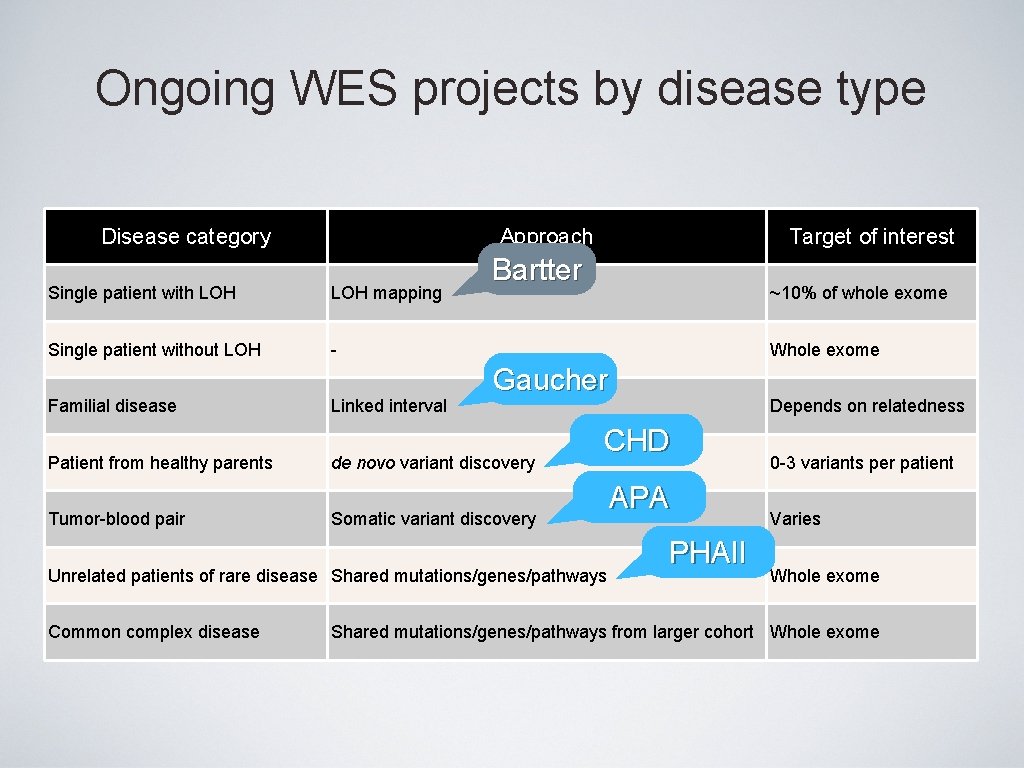
Ongoing WES projects by disease type Disease category Approach Single patient with LOH mapping Single patient without LOH - Familial disease Patient from healthy parents Tumor-blood pair Linked interval Target of interest Bartter ~10% of whole exome Whole exome Gaucher de novo variant discovery CHD Somatic variant discovery Unrelated patients of rare disease Shared mutations/genes/pathways Common complex disease Depends on relatedness APA PHAII 0 -3 variants per patient Varies Whole exome Shared mutations/genes/pathways from larger cohort Whole exome

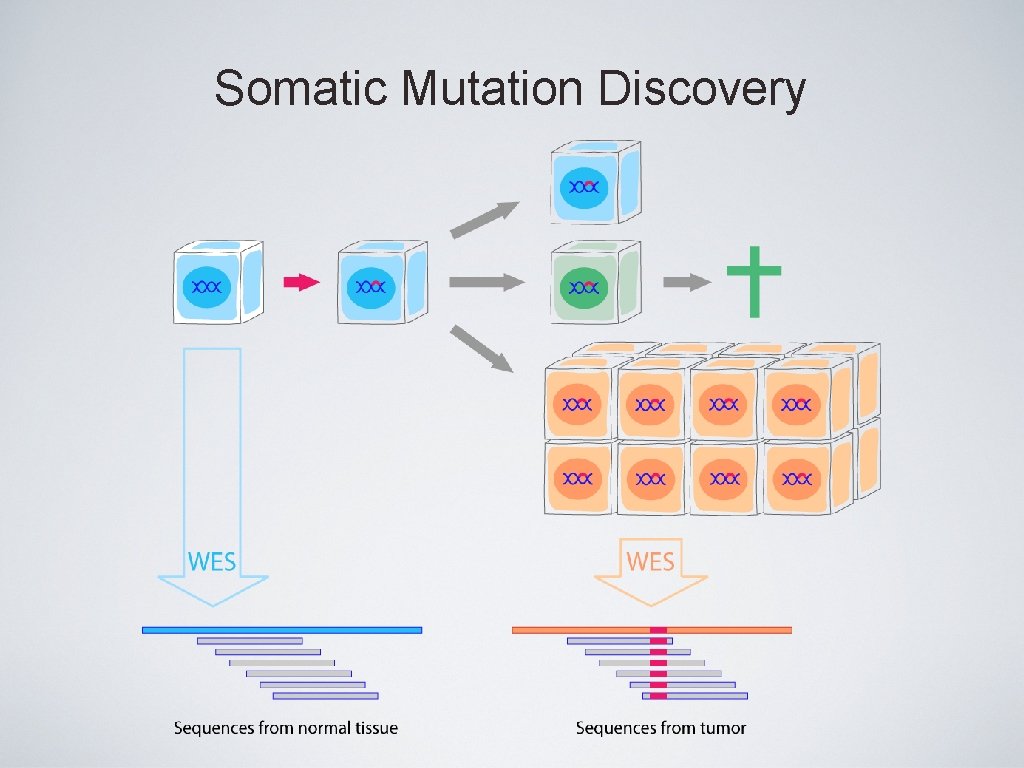
Somatic Mutation Discovery

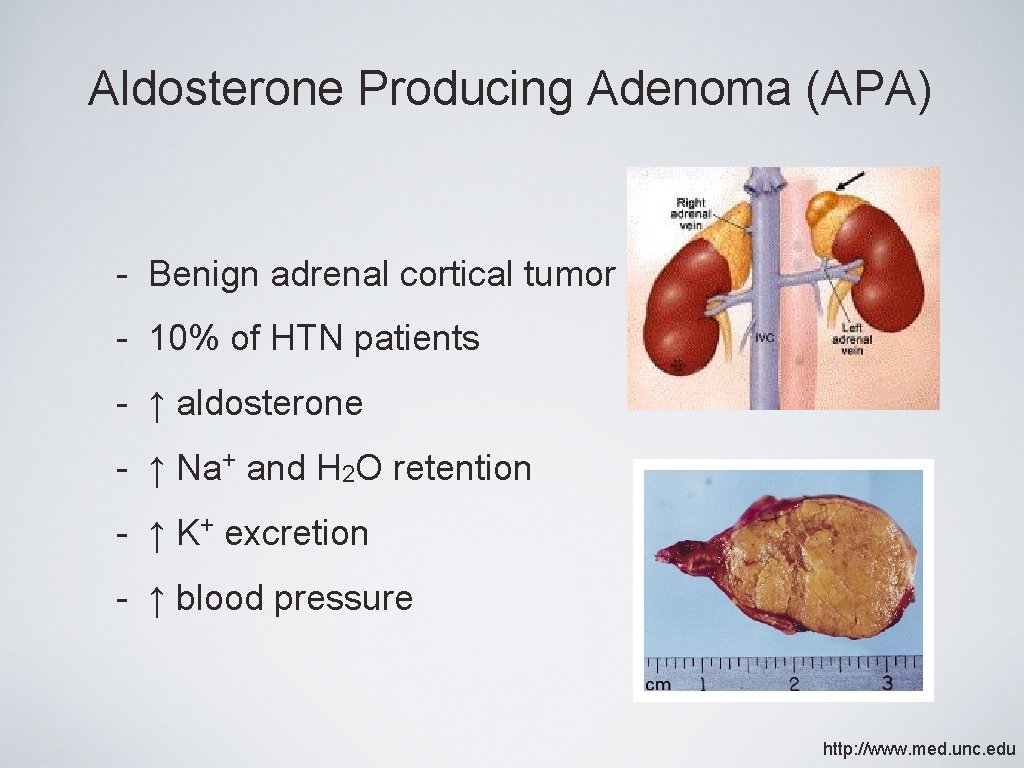
Aldosterone Producing Adenoma (APA) - Benign adrenal cortical tumor - 10% of HTN patients - ↑ aldosterone - ↑ Na+ and H 2 O retention - ↑ K+ excretion - ↑ blood pressure http: //www. med. unc. edu

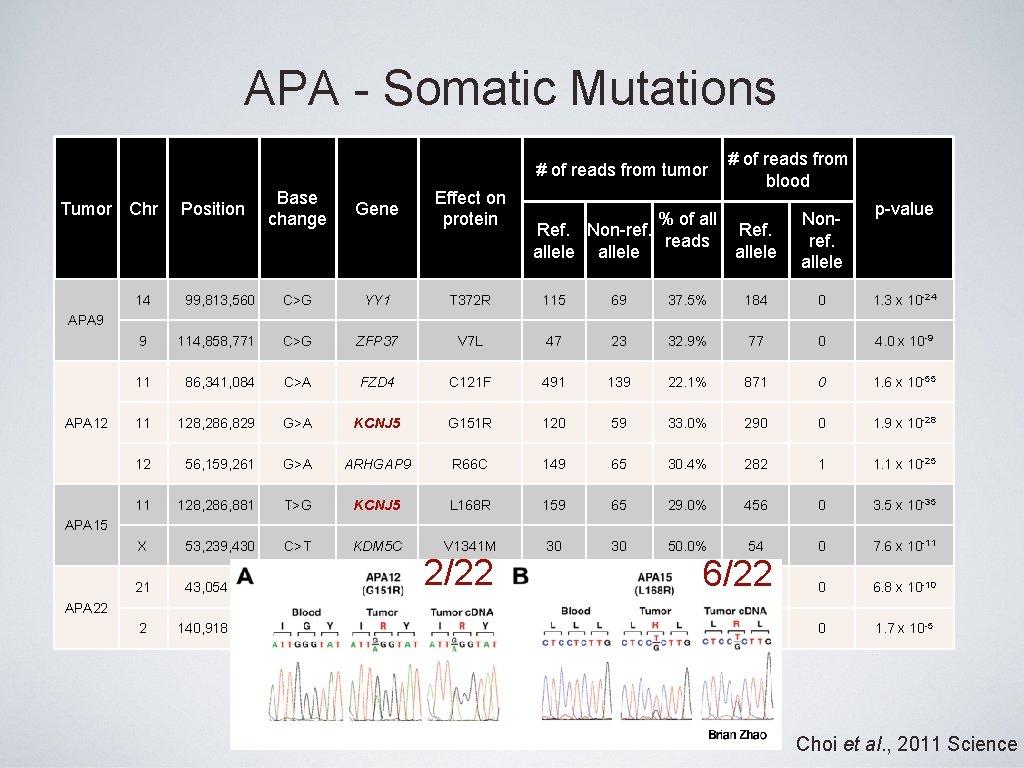
APA - Somatic Mutations # of reads from tumor Tumor Chr Position Base change Gene Effect on protein # of reads from blood % of all Ref. Non-ref. Ref. reads allele Nonref. allele p-value 14 99, 813, 560 C>G YY 1 T 372 R 115 69 37. 5% 184 0 1. 3 x 10 -24 9 114, 858, 771 C>G ZFP 37 V 7 L 47 23 32. 9% 77 0 4. 0 x 10 -9 11 86, 341, 084 C>A FZD 4 C 121 F 491 139 22. 1% 871 0 1. 6 x 10 -55 11 128, 286, 829 G>A KCNJ 5 G 151 R 120 59 33. 0% 290 0 1. 9 x 10 -28 12 56, 159, 261 G>A ARHGAP 9 R 66 C 149 65 30. 4% 282 1 1. 1 x 10 -25 11 128, 286, 881 T>G KCNJ 5 L 168 R 159 65 29. 0% 456 0 3. 5 x 10 -35 X 53, 239, 430 C>T KDM 5 C V 1341 M 30 50. 0% 54 0 7. 6 x 10 -11 21 43, 054, 087 G>A PDE 9 A 2/22 30 Exon 13 splice donor GT>AT 90 31 25. 6% 123 0 6. 8 x 10 -10 2 140, 918, 376 T>G LRP 1 B R 3429 S 60 14 18. 9% 80 0 1. 7 x 10 -5 APA 9 APA 12 APA 15 APA 22 6/22 Choi et al. , 2011 Science

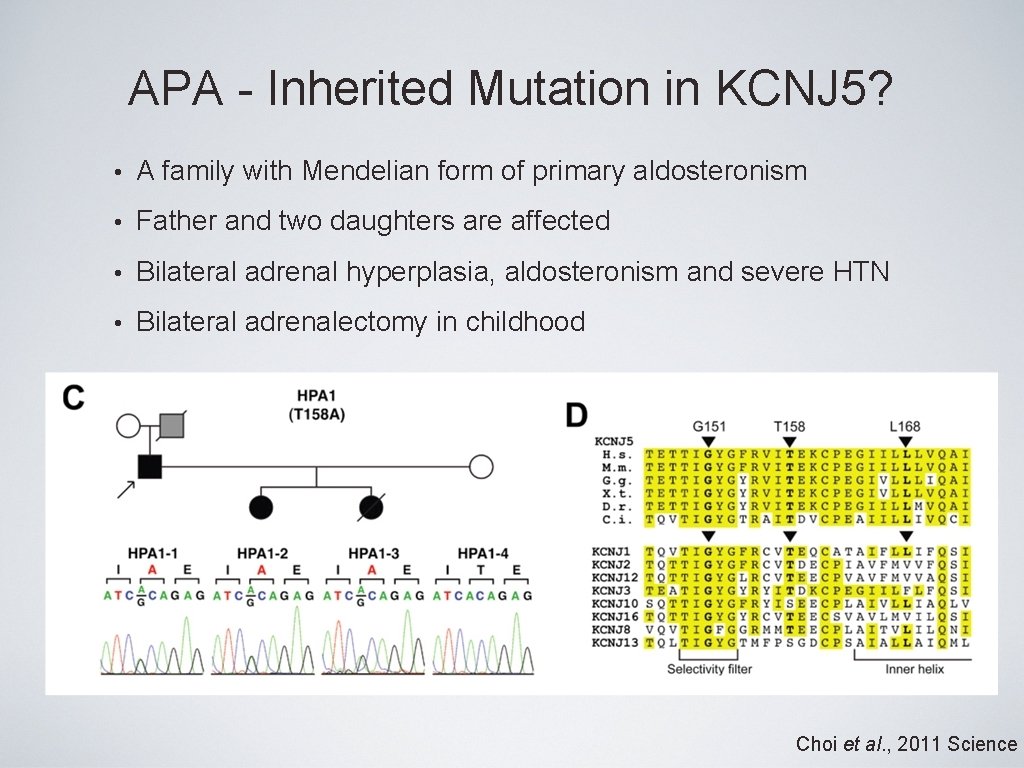
APA - Inherited Mutation in KCNJ 5? • A family with Mendelian form of primary aldosteronism • Father and two daughters are affected • Bilateral adrenal hyperplasia, aldosteronism and severe HTN • Bilateral adrenalectomy in childhood Choi et al. , 2011 Science

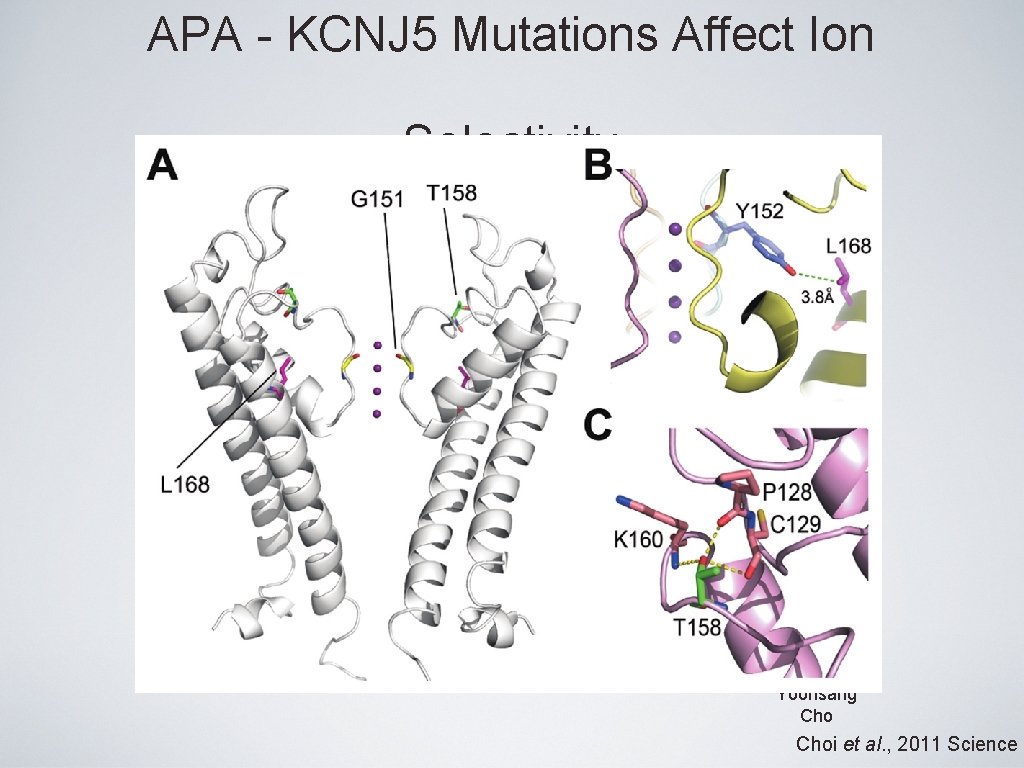
APA - KCNJ 5 Mutations Affect Ion Selectivity Yoonsang Choi et al. , 2011 Science

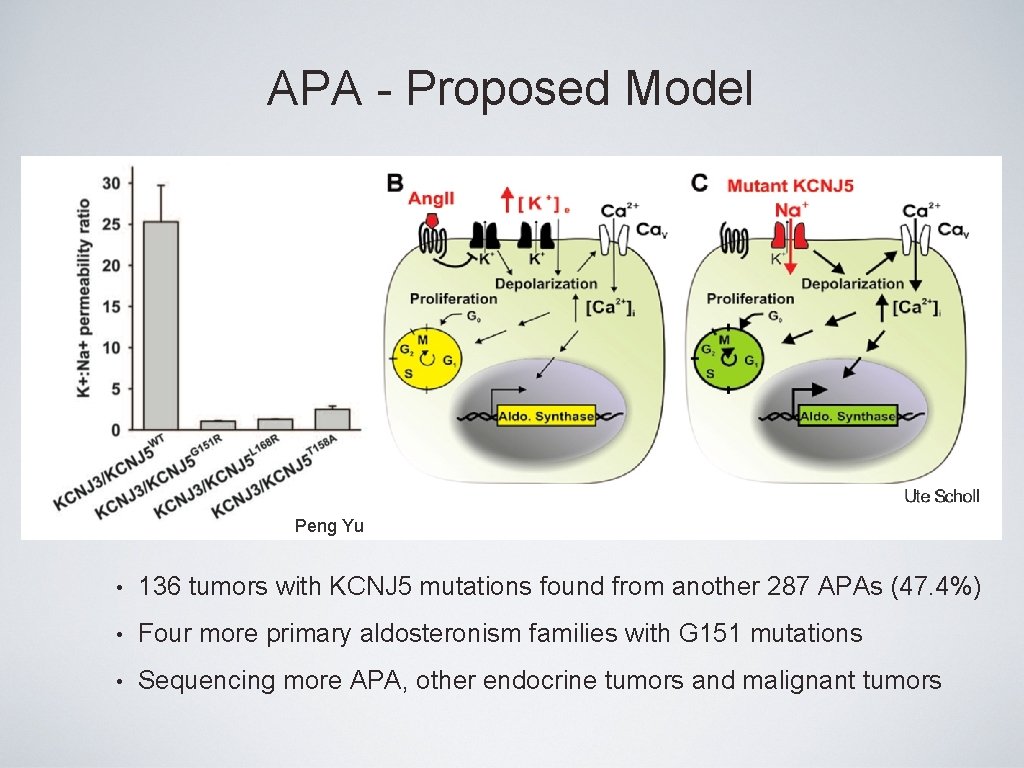
APA - Proposed Model A Peng Yu • 136 tumors with KCNJ 5 mutations found from another 287 APAs (47. 4%) • Four more primary aldosteronism families with G 151 mutations • Sequencing more APA, other endocrine tumors and malignant tumors

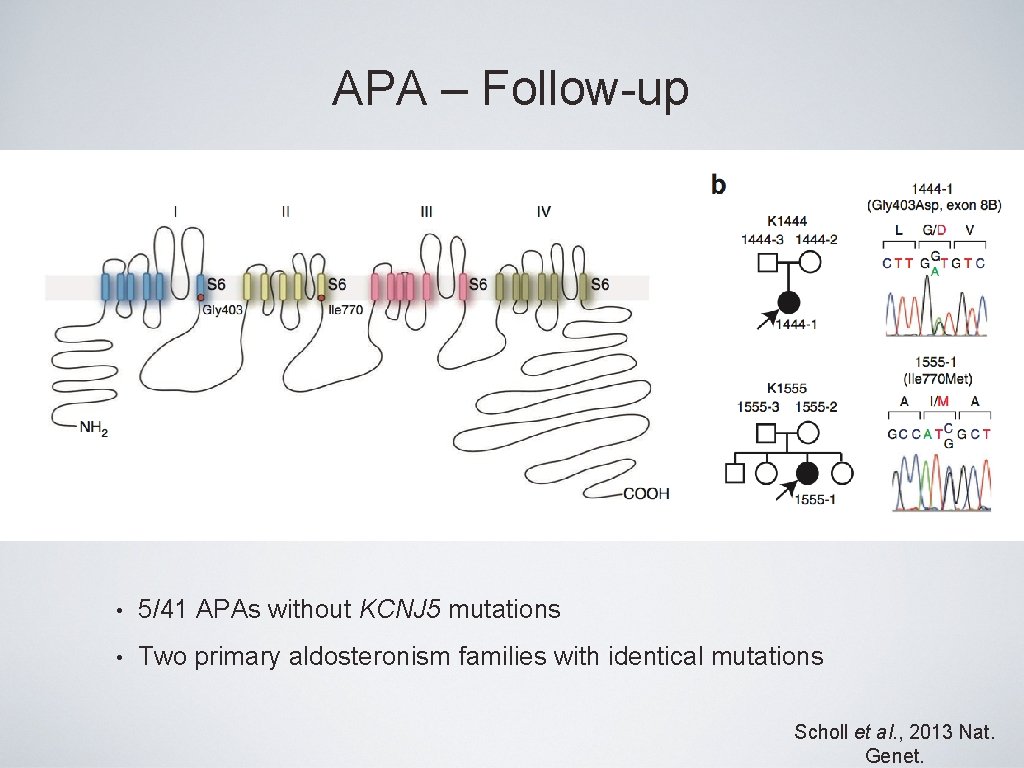
APA – Follow-up • 5/41 APAs without KCNJ 5 mutations • Two primary aldosteronism families with identical mutations Scholl et al. , 2013 Nat. Genet.

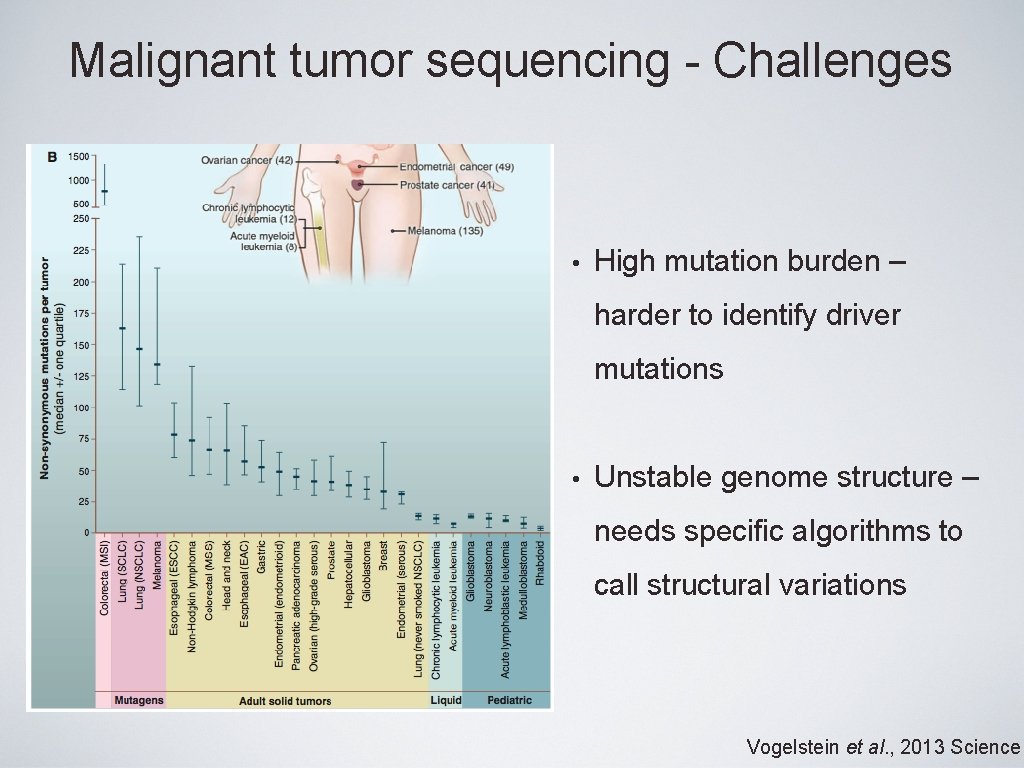
Malignant tumor sequencing - Challenges • High mutation burden – harder to identify driver mutations • Unstable genome structure – needs specific algorithms to call structural variations Vogelstein et al. , 2013 Science

Cancer Sequencing Project at Yale from 2011 • Phase 1: $40 M for 4 years • Phase 2: $60 M for 6 years • PI: Dr. Joseph Schlessinger (Pharmacology) On the scientific committee: Dr. Richard Lifton (Genetics) Dr. Thomas Lynch (Cancer Center) Dr. Roy Herbst (Medical Oncology) Immediate goal: Sequence all cancer samples in Yale’s

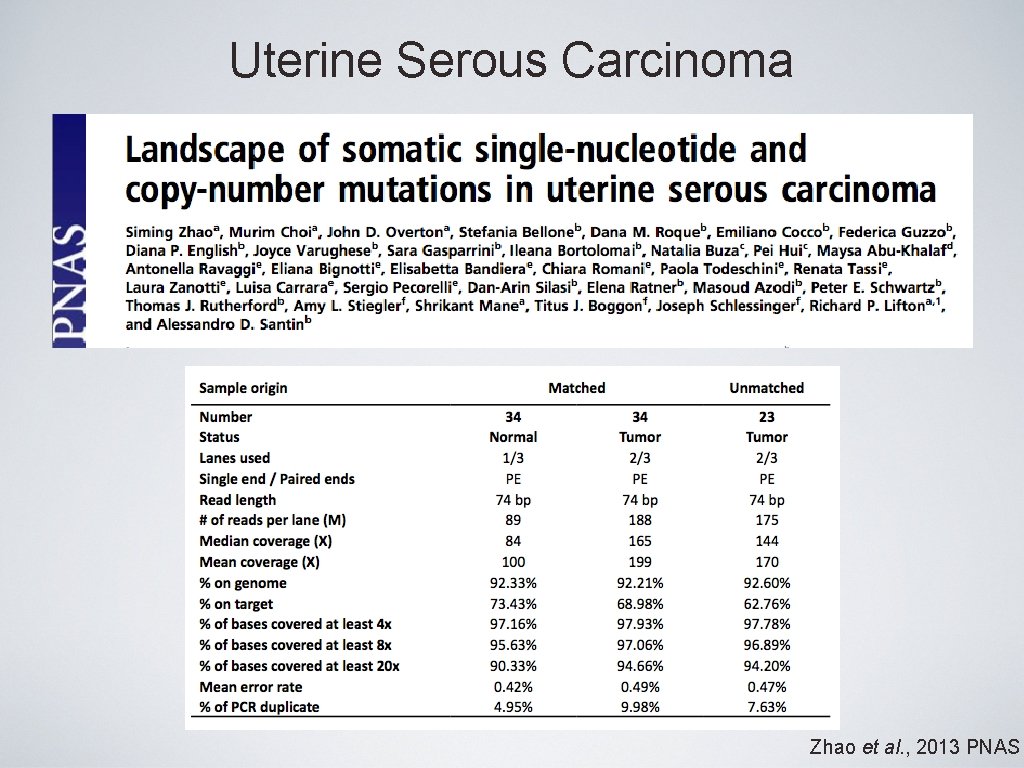
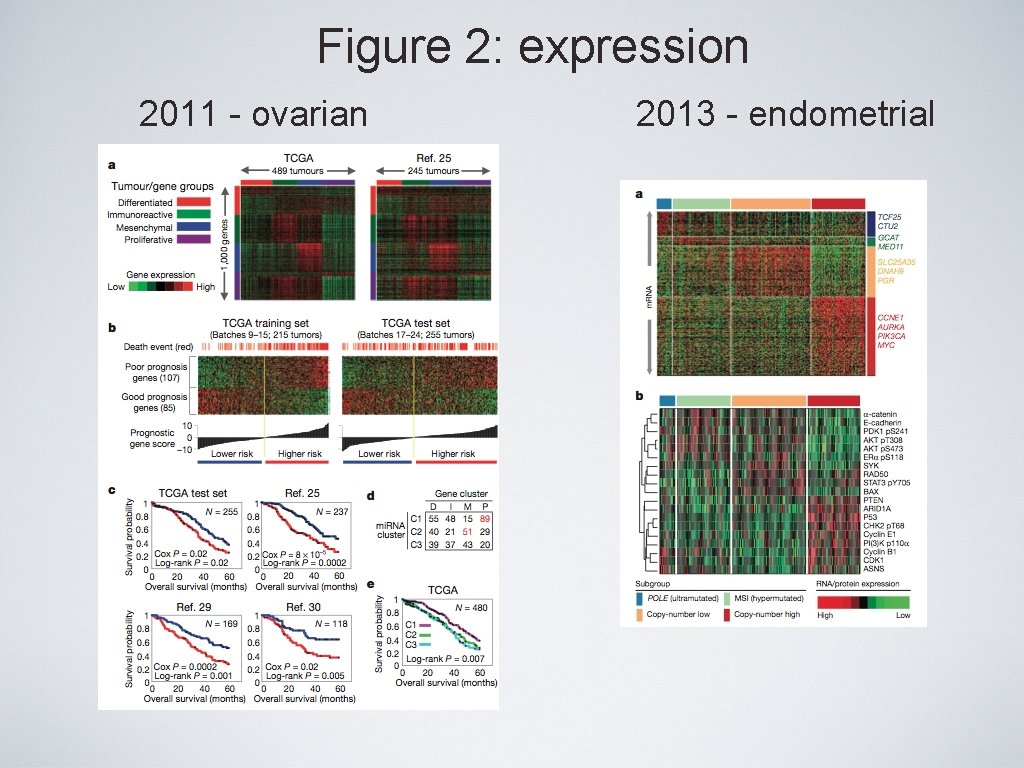
Uterine Serous Carcinoma Zhao et al. , 2013 PNAS

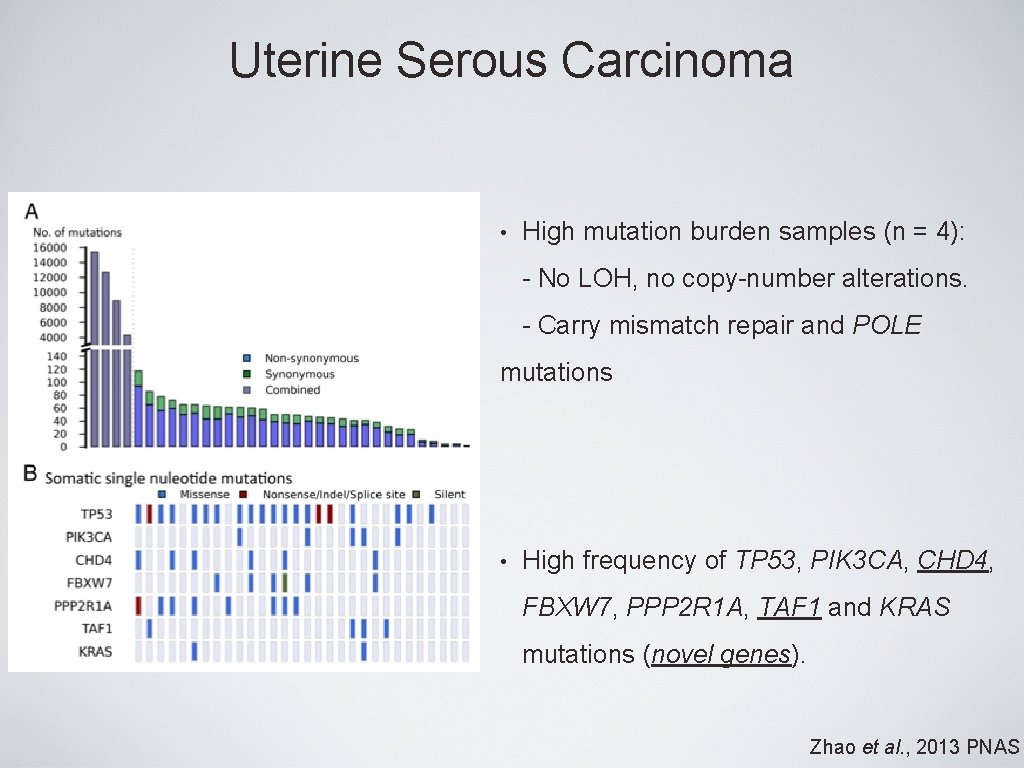
Uterine Serous Carcinoma • High mutation burden samples (n = 4): - No LOH, no copy-number alterations. - Carry mismatch repair and POLE mutations • High frequency of TP 53, PIK 3 CA, CHD 4, FBXW 7, PPP 2 R 1 A, TAF 1 and KRAS mutations (novel genes). Zhao et al. , 2013 PNAS

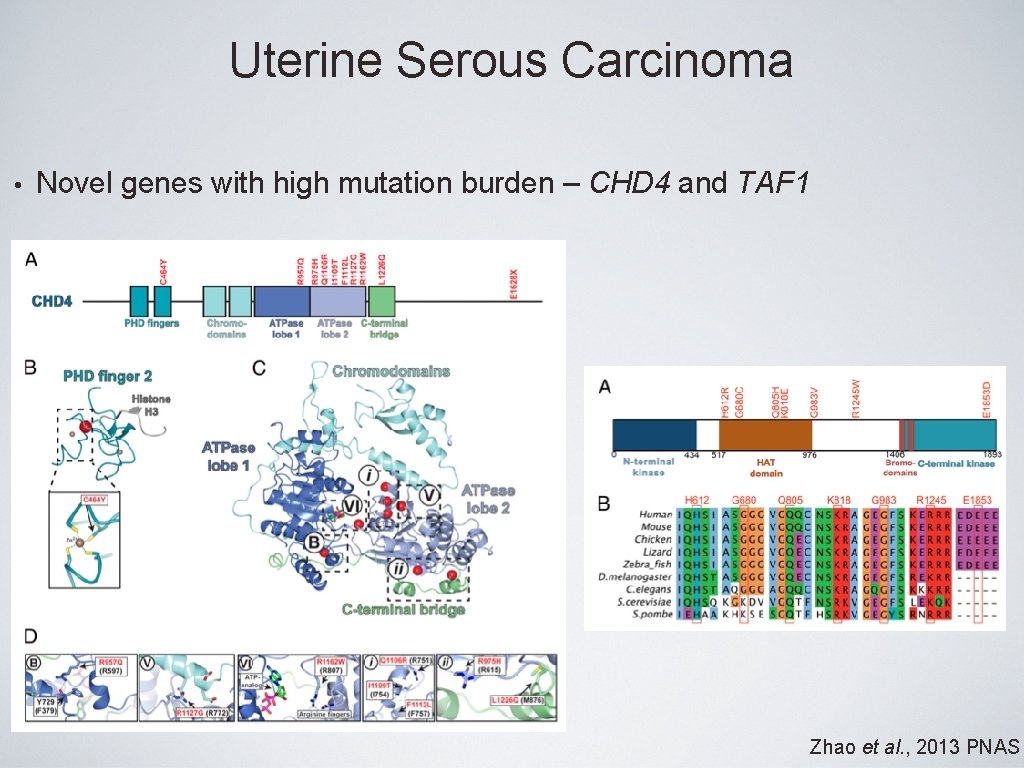
Uterine Serous Carcinoma • Novel genes with high mutation burden – CHD 4 and TAF 1 Zhao et al. , 2013 PNAS

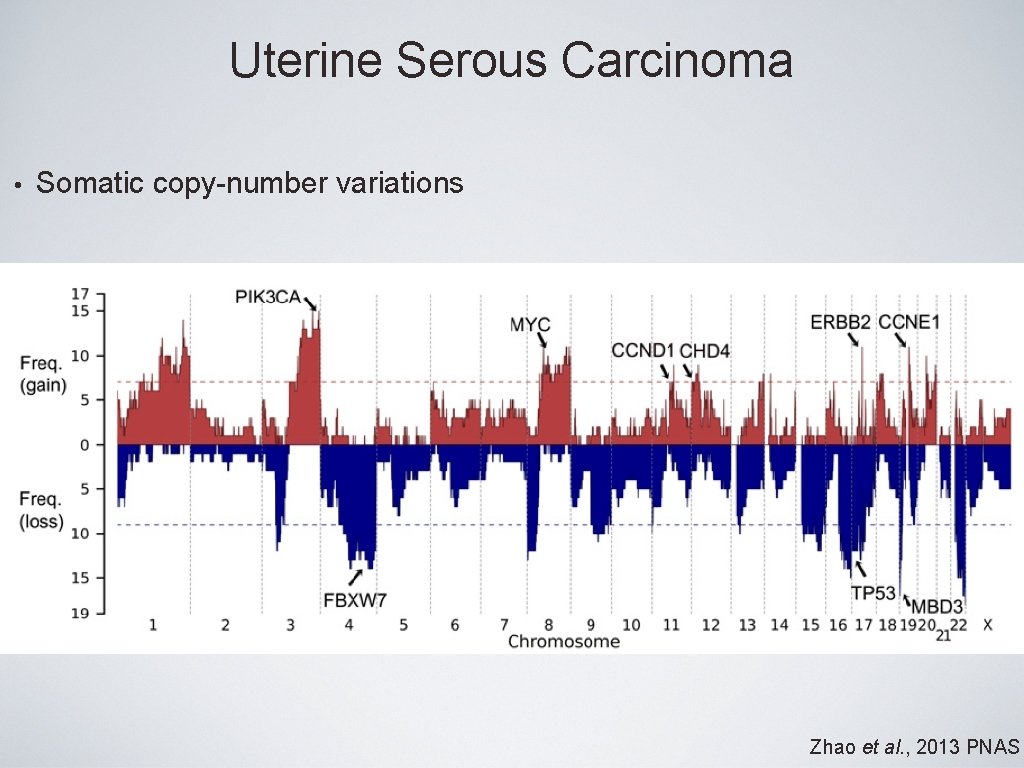
Uterine Serous Carcinoma • Somatic copy-number variations Zhao et al. , 2013 PNAS

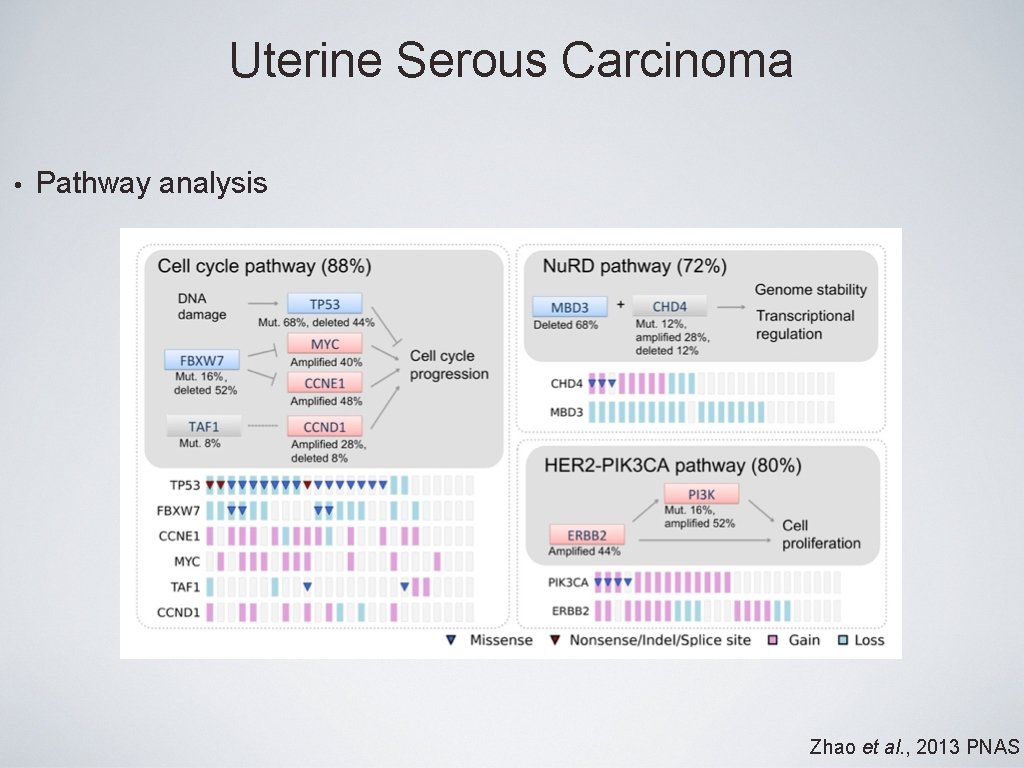
Uterine Serous Carcinoma • Pathway analysis Zhao et al. , 2013 PNAS

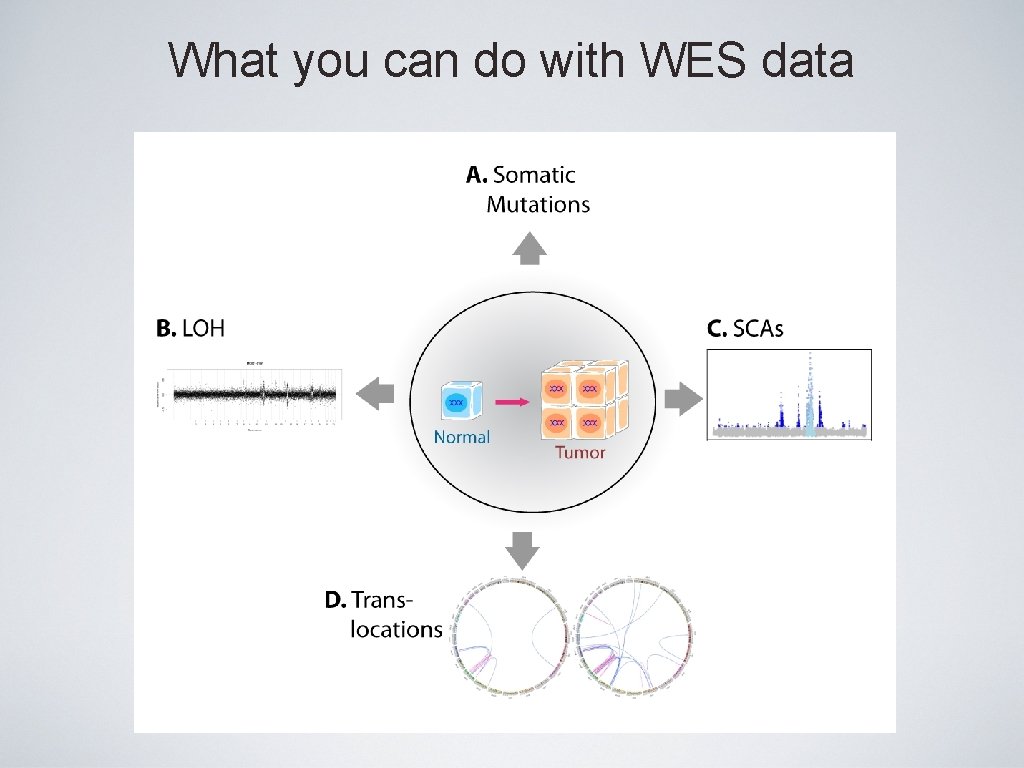
What you can do with WES data

Other Ongoing Cancer Sequencing Projects • Lung ADC (n = 110) + brain met (n = 42) • Lung SCC (n = 110) • Colon cancer with matched metastasis tissues (n = 50) • Others

Prospective trends of cancer genomics 2013 • Large sample set • Data generated by multiple centers and platforms • - Clear cell renal cell carcinoma. Nature - Endometrial carcinoma. Nature - Acute myeloid leukemia. NEJM 2012 - Colorectal cancer. Nature - Squamous cell lung cancer. Nature - Breast cancer. Nature Combines genome, transcriptome, methylome etc 2011 • Leads analysis pipelines - Ovarian cancer. Nature 2008 - Glioblasoma. Nature

2011, n = 489 2013, n = 363

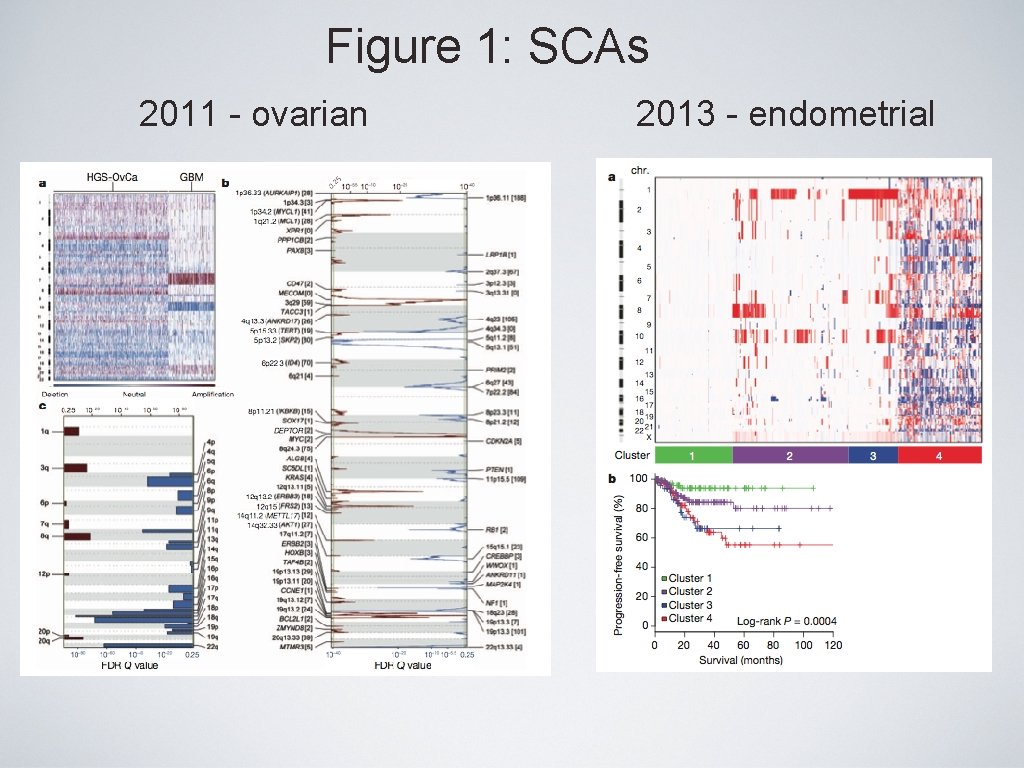
Figure 1: SCAs 2011 - ovarian 2013 - endometrial

Figure 2: expression 2011 - ovarian 2013 - endometrial

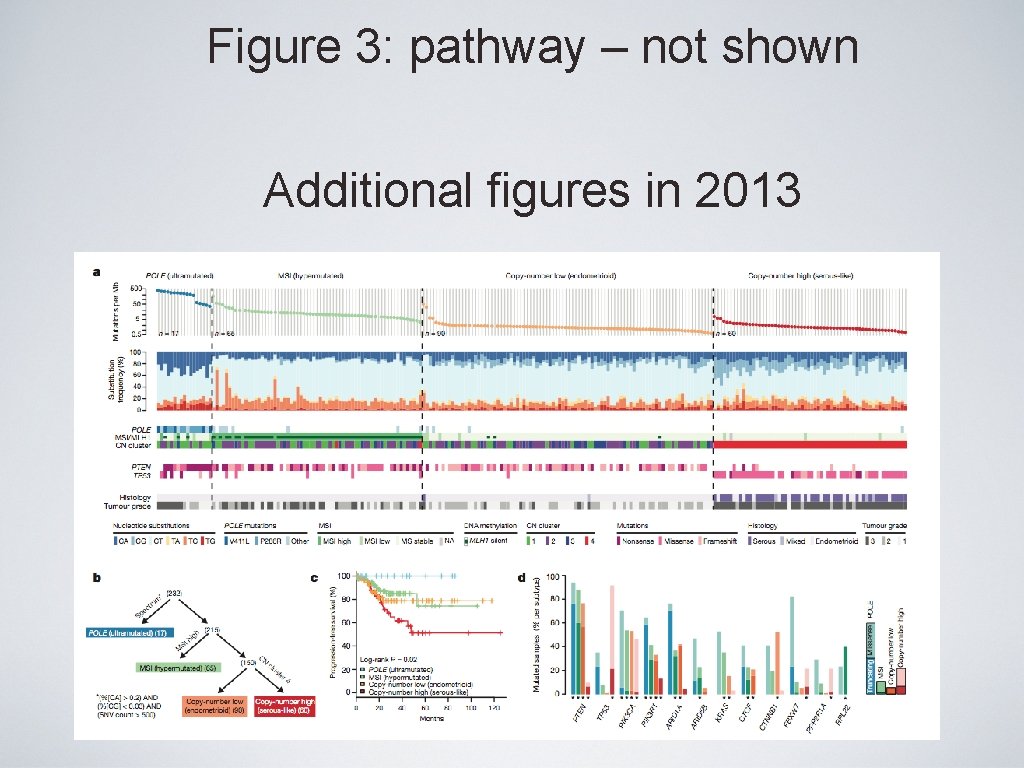
Figure 3: pathway – not shown Additional figures in 2013

Predicting the future • Increasing sample size • Combining multiple omics data + clinical data • Metastasis, recurrence problem • Multi-clonality problem • Circulating tumor DNA

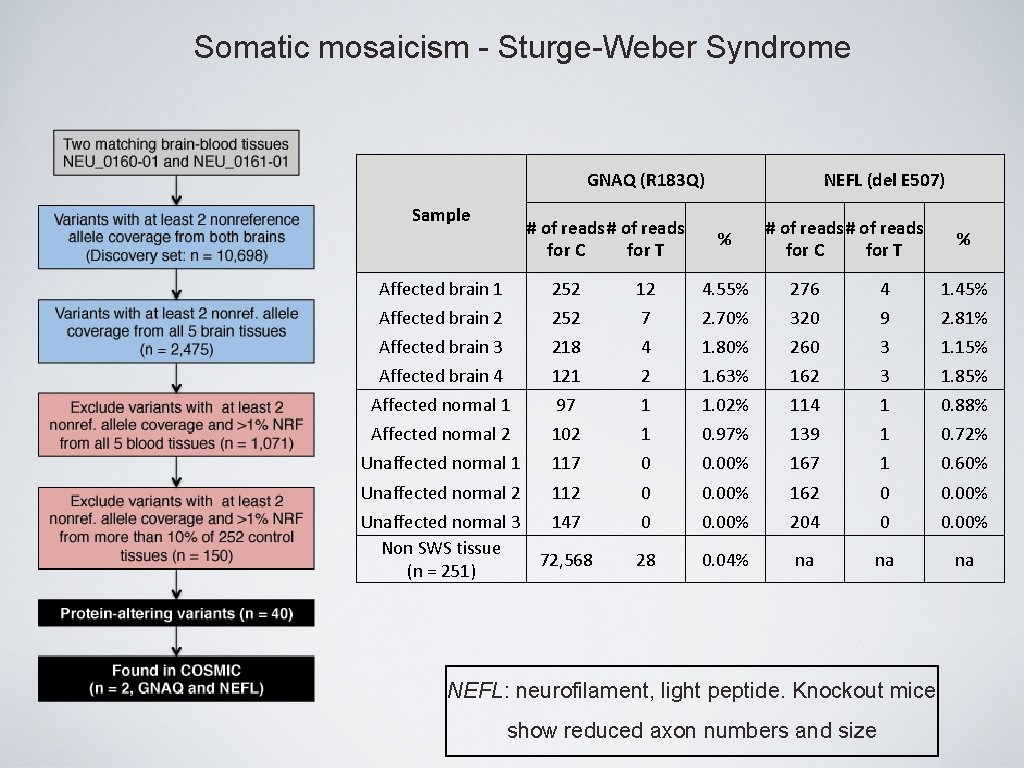
Somatic mosaicism - Sturge-Weber Syndrome • Cohort: WES of brain tissues from 4 affected subjects • Cohort: WGS of matched brain and blood tissues from 3 affected subjects + 50 subjects for replication • Result: Somatic GNAQ Arg 183 Gln change with very low % (1 -18%) Shirley et al. , 2013 NEJM

Somatic mosaicism - Sturge-Weber Syndrome GNAQ (R 183 Q) Sample # of reads for C for T NEFL (del E 507) % # of reads for C for T % Affected brain 1 252 12 4. 55% 276 4 1. 45% Affected brain 2 252 7 2. 70% 320 9 2. 81% Affected brain 3 218 4 1. 80% 260 3 1. 15% Affected brain 4 121 2 1. 63% 162 3 1. 85% Affected normal 1 97 1 1. 02% 114 1 0. 88% Affected normal 2 102 1 0. 97% 139 1 0. 72% Unaffected normal 1 117 0 0. 00% 167 1 0. 60% Unaffected normal 2 112 0 0. 00% 162 0 0. 00% 204 0 0. 00% 28 0. 04% na na na Unaffected normal 3 147 Non SWS tissue 72, 568 (n = 251) NEFL: neurofilament, light peptide. Knockout mice show reduced axon numbers and size

Conclusion 1. Whole exome sequencing is proved to be a powerful tool for disease-causing variant discovery 2. WES can be applied to various types of human diseases for (a) clinical diagnosis (b) understanding molecular mechanism

Acknowledgements CLD Ute Scholl Weizhen Ji Lifton lab, HHMI Richard Lifton APA Ute Scholl Tobias Carling Peng Yue (NY Med) Yoonsang Cho (Saint Louis Univ) Brian Zhao CHD Samir Zaidi Martina Brueckner PHAII Lynn Boyden Kaya Bilguvar Murat Gunel Stephan Sanders Matthew State Gaucher Sarah Lo Pramod Mistry Yale Center for Genome Analysis Shrikant Mane John Overton Irina Tikhonova Alex Lopez Computer Science Robert Bjornson Nicholas Carriero http: //genomics. snu. ac. kr Yongjin Yoo Youngha Lee Hyosuk Cho UCSD Yoonsung Lee