Efficient Algorithms for Locating the Length Constrained Heaviest


![Applications to Biomolecular Sequence Analysis (II) • Annotating Multiple Sequence Alignments – [Stojanovic’ 99]: Applications to Biomolecular Sequence Analysis (II) • Annotating Multiple Sequence Alignments – [Stojanovic’ 99]:](https://slidetodoc.com/presentation_image/22ced054ef60a50b2cc5e4e8aedf5038/image-6.jpg)





- Slides: 27

Efficient Algorithms for Locating the Length. Constrained Heaviest Segments, with Applications to Biomolecular Sequence Analysis Yaw-Ling Lin* Tao Jiang Kun-Mao Chao Dept CS & Info Mngmt, Providence Univ, Taiwan Dept CS & Engineering, UC Riverside, USA Dept CS & Info Engnr, Nat. Taiwan Univ, Taiwan * Yaw-Ling Lin, Providence, Taiwan

Outline • • • Introduction. Applications to Biomolecular Sequence Analysis. Maximum Sum Consecutive Subsequence. Maximum Average Consecutive Subsequence. Implementation and Preliminary Experiments Concluding Remarks Yaw-Ling Lin, Providence, Taiwan 2

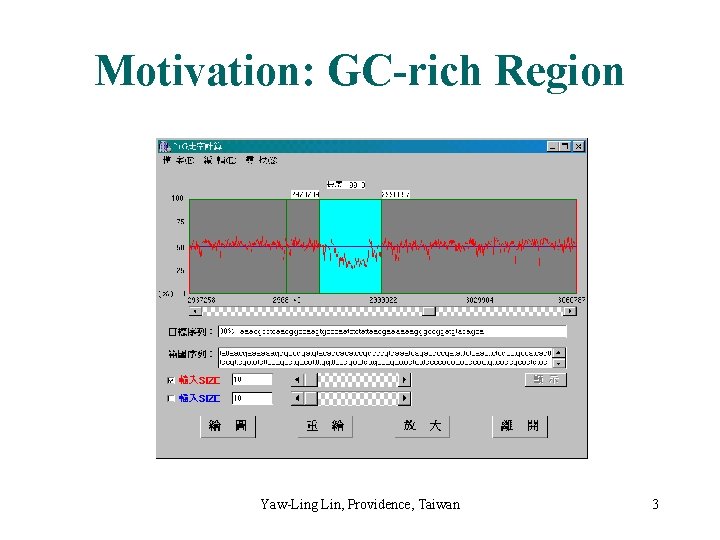
Motivation: GC-rich Region Yaw-Ling Lin, Providence, Taiwan 3

Introduction • Two fundamental algorithms in searching for interesting regions in sequences: • Given a sequence of real numbers of length n and an upper bound U, find a consecutive subsequence of length at most U with the maximum sum --- an O(n)-time algorithm. • Given a sequence of real numbers of length n and a lower bound L, find a consecutive subsequence of length at least L with the maximum average. --- an O(n log L)-time algorithm. Yaw-Ling Lin, Providence, Taiwan 4

Applications to Biomolecular Sequence Analysis (I) • Locating GC-Rich Regions – Finding GC-rich regions: an important problem in gene recognition and comparative genomics. – Cp. G islands ( 200 ~ 1400 bp ) – [Huang’ 94]: O(n L)-time algorithm. • Post-Processing Sequence Alignments – Comparative analysis of human and mouse DNA: useful in gene prediction in human genome. – Mosaic effect: bad inner sequence. – Normalized local alignment. – Post-processing local aligned subsequences Yaw-Ling Lin, Providence, Taiwan 5
![Applications to Biomolecular Sequence Analysis II Annotating Multiple Sequence Alignments Stojanovic 99 Applications to Biomolecular Sequence Analysis (II) • Annotating Multiple Sequence Alignments – [Stojanovic’ 99]:](https://slidetodoc.com/presentation_image/22ced054ef60a50b2cc5e4e8aedf5038/image-6.jpg)
Applications to Biomolecular Sequence Analysis (II) • Annotating Multiple Sequence Alignments – [Stojanovic’ 99]: conserved regions in biomolecular sequences. – Numerical scores for columns of a multiple alignment; each column score shall be adjusted by subtracting an anchor value. • Ungapped Local Alignments with Length Constraints – Computing the length-constrained segment of each diagonal in the matrix with the largest sum (or average) of scores. – Applications in motif identification. Yaw-Ling Lin, Providence, Taiwan 6

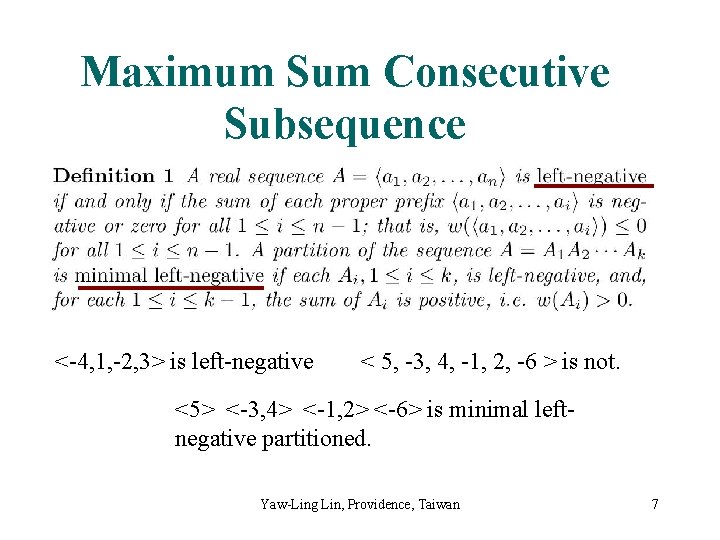
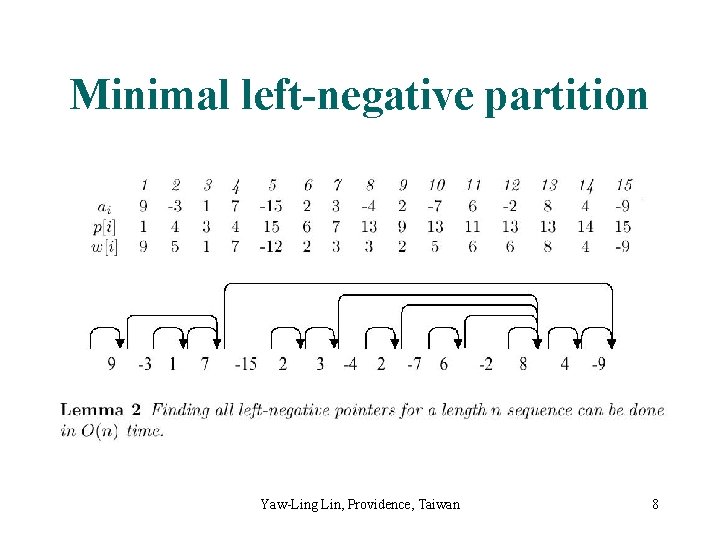
Maximum Sum Consecutive Subsequence <-4, 1, -2, 3> is left-negative < 5, -3, 4, -1, 2, -6 > is not. <5> <-3, 4> <-1, 2> <-6> is minimal leftnegative partitioned. Yaw-Ling Lin, Providence, Taiwan 7

Minimal left-negative partition Yaw-Ling Lin, Providence, Taiwan 8

MLN-partition: linear time Yaw-Ling Lin, Providence, Taiwan 9

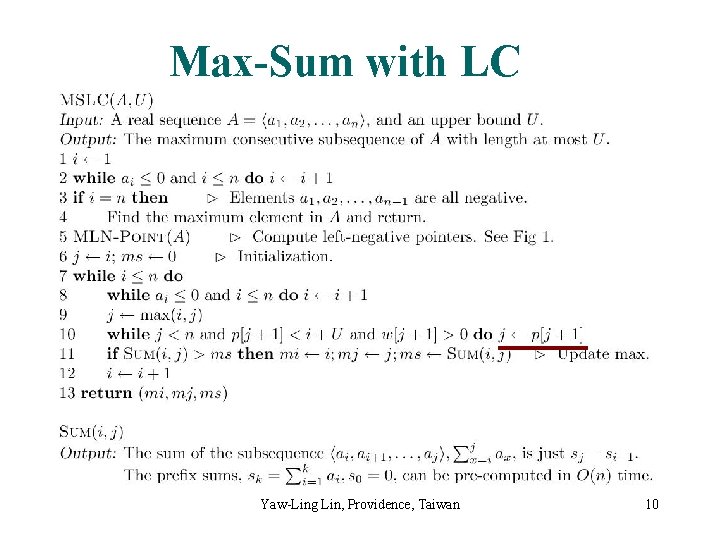
Max-Sum with LC Yaw-Ling Lin, Providence, Taiwan 10

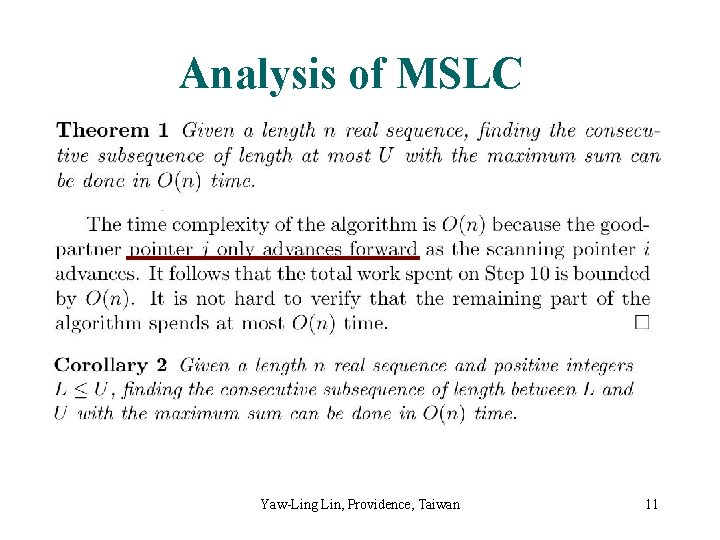
Analysis of MSLC Yaw-Ling Lin, Providence, Taiwan 11

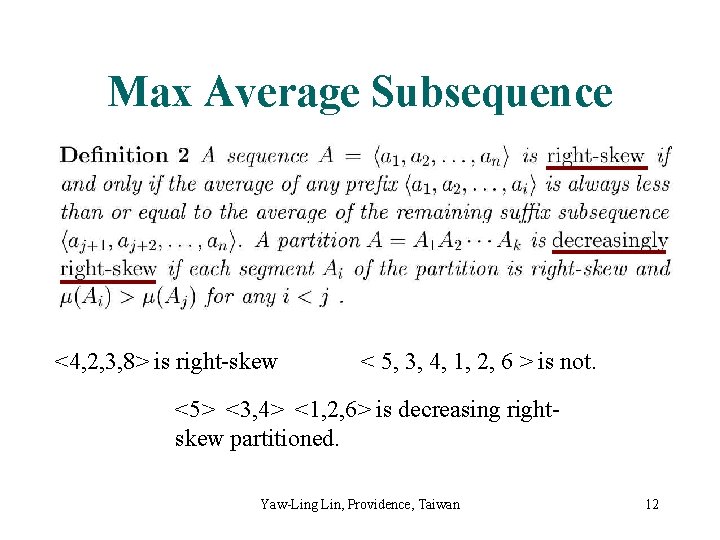
Max Average Subsequence <4, 2, 3, 8> is right-skew < 5, 3, 4, 1, 2, 6 > is not. <5> <3, 4> <1, 2, 6> is decreasing rightskew partitioned. Yaw-Ling Lin, Providence, Taiwan 12

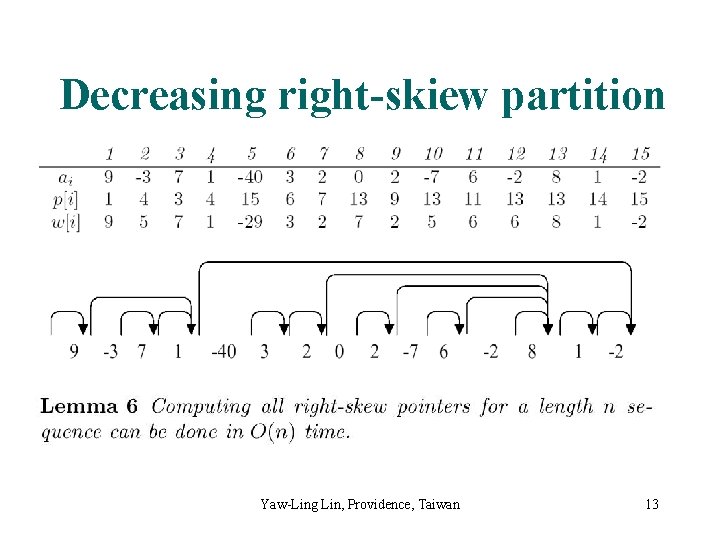
Decreasing right-skiew partition Yaw-Ling Lin, Providence, Taiwan 13

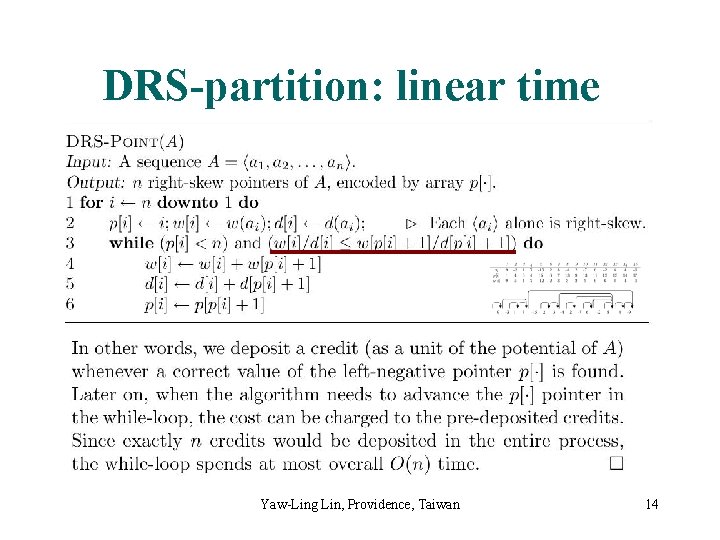
DRS-partition: linear time Yaw-Ling Lin, Providence, Taiwan 14

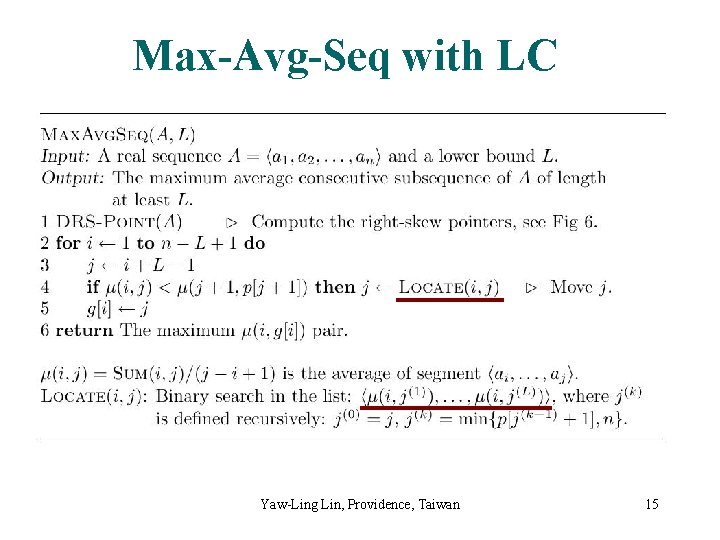
Max-Avg-Seq with LC Yaw-Ling Lin, Providence, Taiwan 15

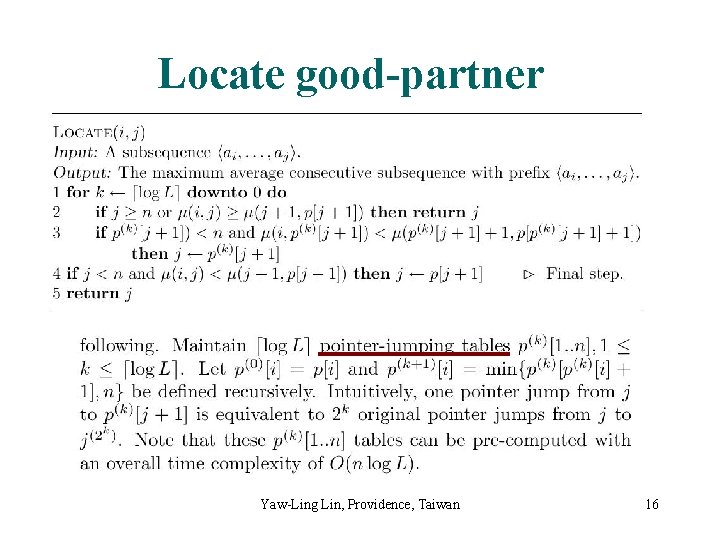
Locate good-partner Yaw-Ling Lin, Providence, Taiwan 16

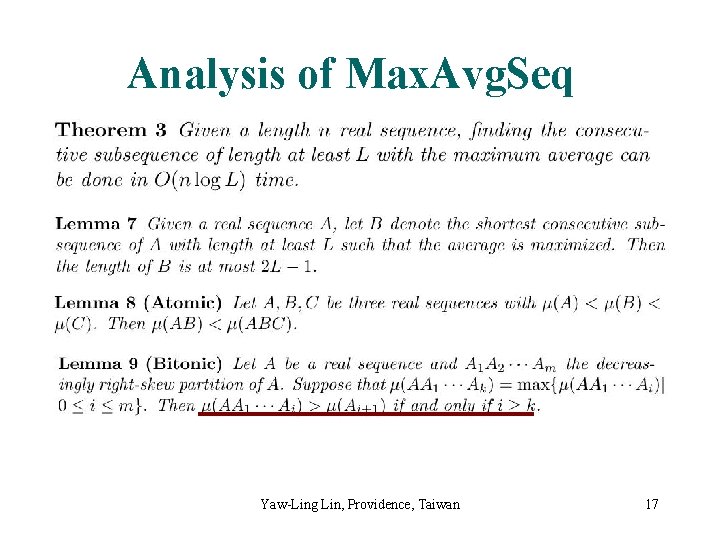
Analysis of Max. Avg. Seq Yaw-Ling Lin, Providence, Taiwan 17

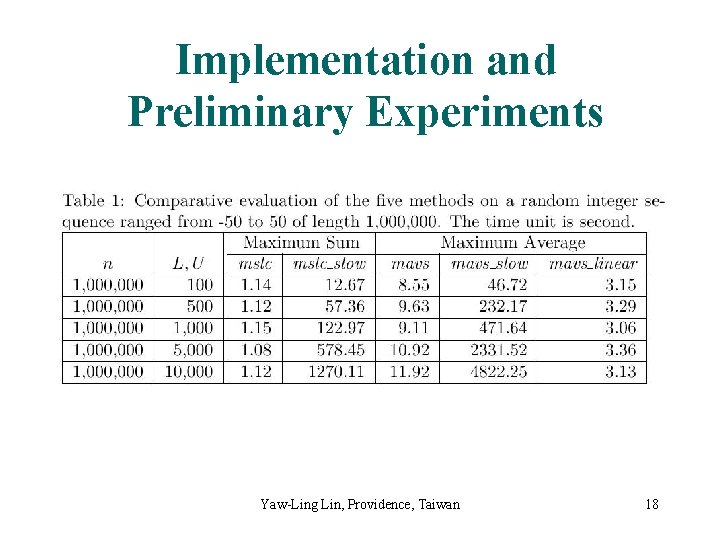
Implementation and Preliminary Experiments Yaw-Ling Lin, Providence, Taiwan 18

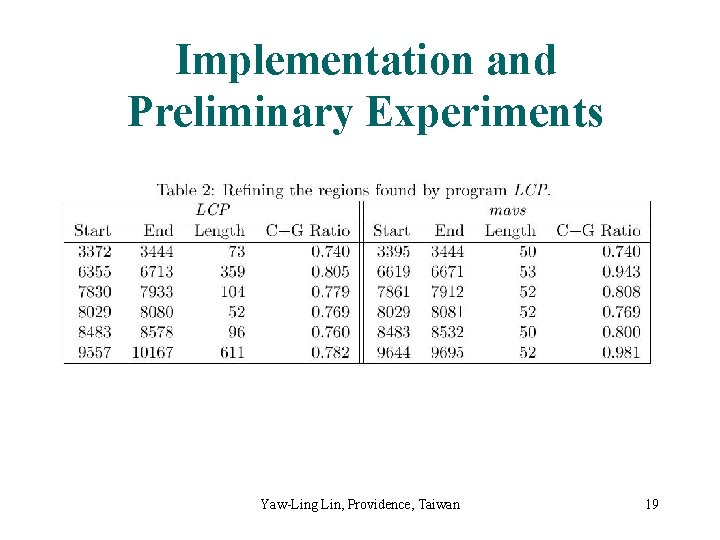
Implementation and Preliminary Experiments Yaw-Ling Lin, Providence, Taiwan 19

Conclusion • Find a max-sum subsequence of length at most U can be done in O(n)-time. • Find a max-avg subsequence of length at least L can be done in O(n log L)-time. Yaw-Ling Lin, Providence, Taiwan 20

Recent Progress • Lu (CMCT’ 2002): finding the max-avg subsequence of length at least L on binary (0, 1) sequences. O(n)-time. • Goldwasser, Kao, Lu (WABI’ 2002): finding the max-avg subsequence of length at least L and at most U on real sequences. O(n)-time • Tools: finding Cp. G islands using MAVG (joint work with Huang, X. , Jiang, T. and Chao, K. -M. ) http: //deepc 2. zool. iastate. edu/aat/mavg/cgdoc. html http: //deepc 2. zool. iastate. edu/aat/mavg/cg. html Yaw-Ling Lin, Providence, Taiwan 21

Goldwasser, Kao, Lu (WABI’ 2002)’s Linear-Time Algorithm Yaw-Ling Lin, Providence, Taiwan

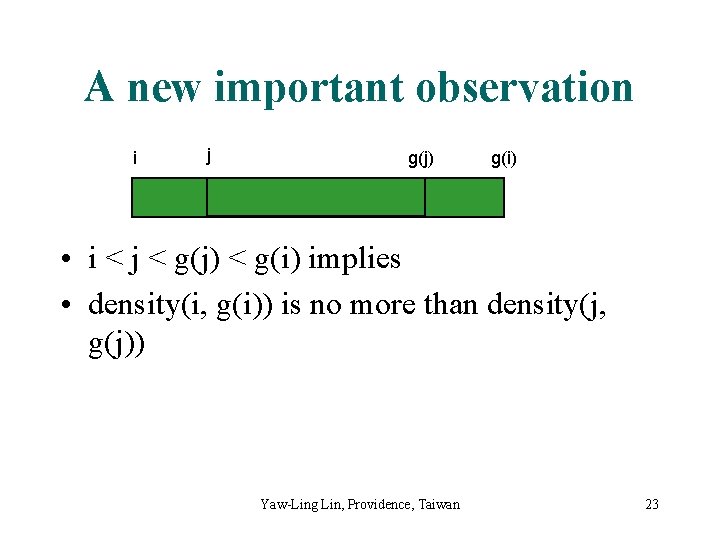
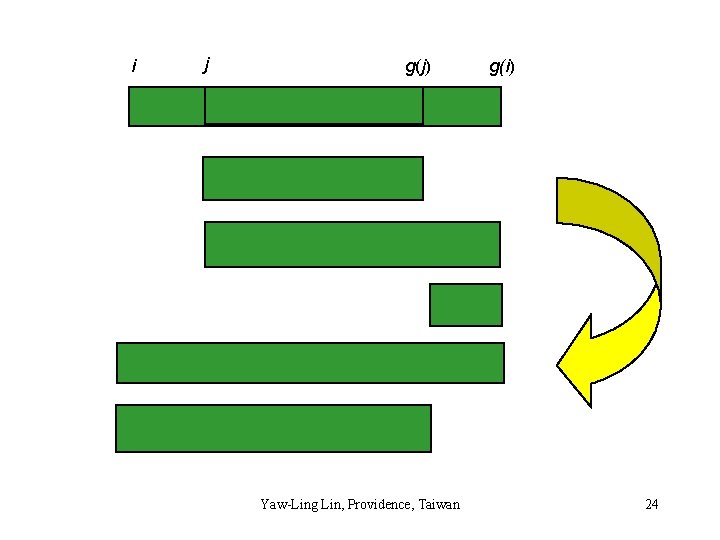
A new important observation i j g(j) g(i) • i < j < g(j) < g(i) implies • density(i, g(i)) is no more than density(j, g(j)) Yaw-Ling Lin, Providence, Taiwan 23

i j g(j) Yaw-Ling Lin, Providence, Taiwan g(i) 24

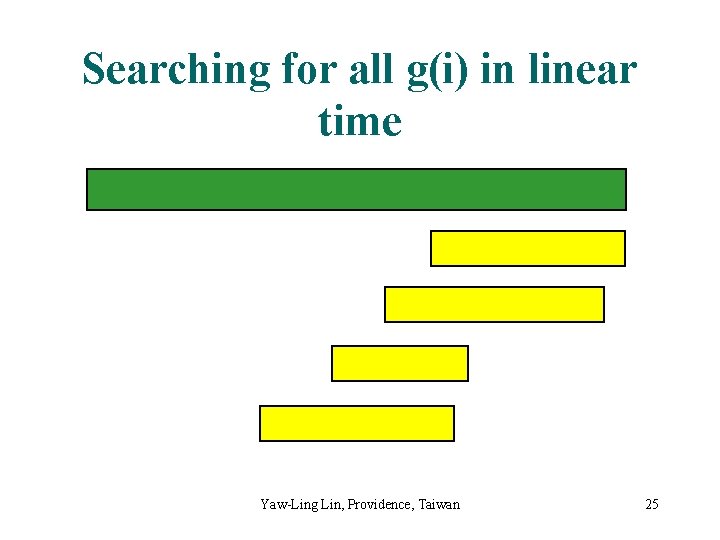
Searching for all g(i) in linear time Yaw-Ling Lin, Providence, Taiwan 25

Some thoughts • Attacking new problems with new ideas. • Collaboration is important for bioinformatics – Communication – Work on what you are good at Yaw-Ling Lin, Providence, Taiwan 26

Future Research • Best k (nonintersecting) subsequences? • Normalized local alignment? • Measurement of goodness? Yaw-Ling Lin, Providence, Taiwan 27