Effective Dimension Reduction with Prior Knowledge Haesun Park



- Slides: 18

Effective Dimension Reduction with Prior Knowledge Haesun Park Division of Computational Science and Eng. College of Computing Georgia Institute of Technology Atlanta, GA Joint work w/ Barry Drake, Peg Howland, Hyunsoo Kim, and Cheonghee Park DIMACS, May, 2007

Dimension Reduction • Dimension Reduction for Clustered Data: Linear Discriminant Analysis (LDA) Generalized LDA (LDA/GSVD, regularized LDA) Orthogonal Centroid Method (OCM) • Dimension Reduction for Nonnegative Data: Nonnegative Matrix Factorization (NMF) • Applications: Text classification, Face recognition, Fingerprint classification, Gene clustering in Microarray Analysis …

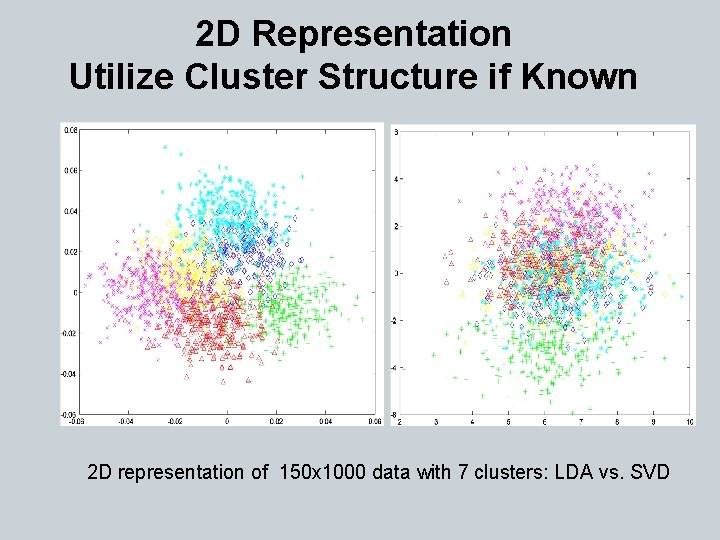
2 D Representation Utilize Cluster Structure if Known 2 D representation of 150 x 1000 data with 7 clusters: LDA vs. SVD

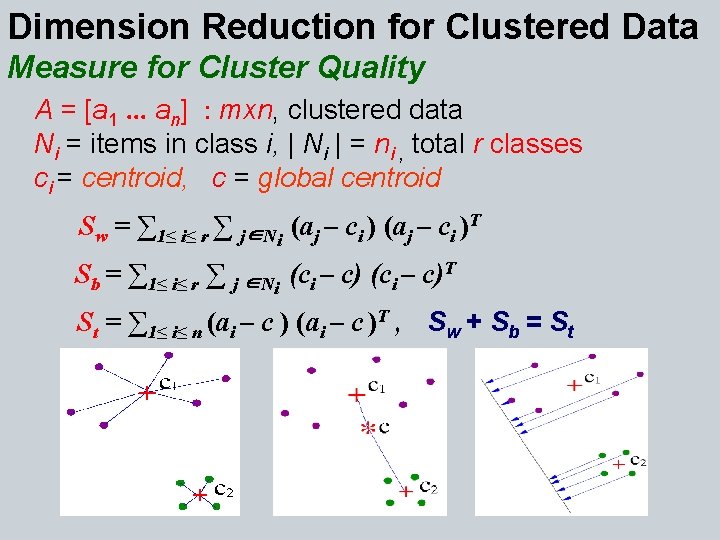
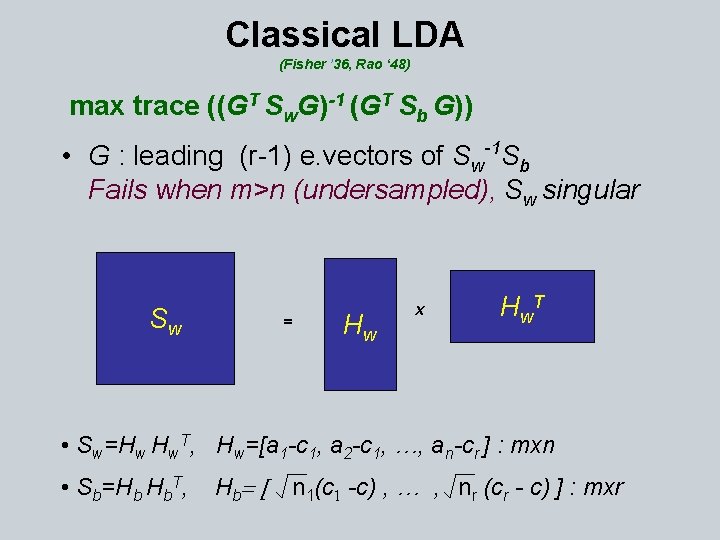
Dimension Reduction for Clustered Data Measure for Cluster Quality A = [a 1. . . an] : mxn, clustered data Ni = items in class i, | Ni | = ni , total r classes ci = centroid, c = global centroid Sw = ∑ 1≤ i≤ r ∑ j∈Ni (aj – ci )T Sb = ∑ 1≤ i≤ r ∑ j ∈Ni (ci – c)T St = ∑ 1≤ i≤ n (ai – c )T , Sw + Sb = St

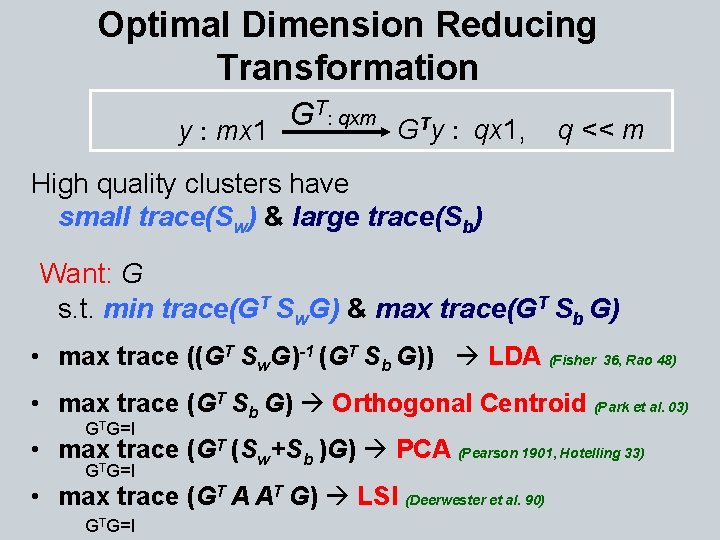
Optimal Dimension Reducing Transformation T: qxm G GTy : qx 1, q << m y : mx 1 High quality clusters have small trace(Sw) & large trace(Sb) Want: G s. t. min trace(GT Sw. G) & max trace(GT Sb G) • max trace ((GT Sw. G)-1 (GT Sb G)) LDA (Fisher 36, Rao 48) • max trace (GT Sb G) Orthogonal Centroid (Park et al. 03) GTG=I • max. T trace (GT (Sw+Sb )G) PCA (Pearson 1901, Hotelling 33) G G=I • max trace (GT A AT G) LSI (Deerwester et al. 90) GTG=I

Classical LDA (Fisher ’ 36, Rao ‘ 48) max trace ((GT Sw. G)-1 (GT Sb G)) • G : leading (r-1) e. vectors of Sw-1 Sb Fails when m>n (undersampled), Sw singular Sw = Hw x H w. T • Sw=Hw Hw. T, Hw=[a 1 -c 1, a 2 -c 1, …, an-cr ] : mxn • Sb=Hb Hb. T, H b= [ n 1(c 1 -c) , … , nr (cr - c) ] : mxr

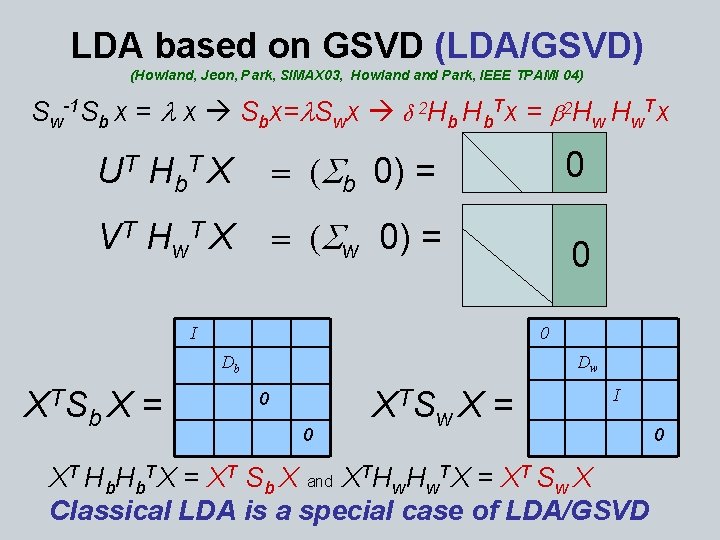
LDA based on GSVD (LDA/GSVD) (Howland, Jeon, Park, SIMAX 03, Howland Park, IEEE TPAMI 04) Sw-1 Sb x = l x Sbx=l. Swx δ 2 Hb Hb. Tx = b 2 Hw Hw. Tx U T H b. T X = (Sb 0) = 0 V T H w. T X = (Sw 0) = 0 0 I Dw Db X TS b. X = X TS 0 0 w. X = I XT Hb. TX = XT Sb X and XTHw. TX = XT Sw X Classical LDA is a special case of LDA/GSVD 0

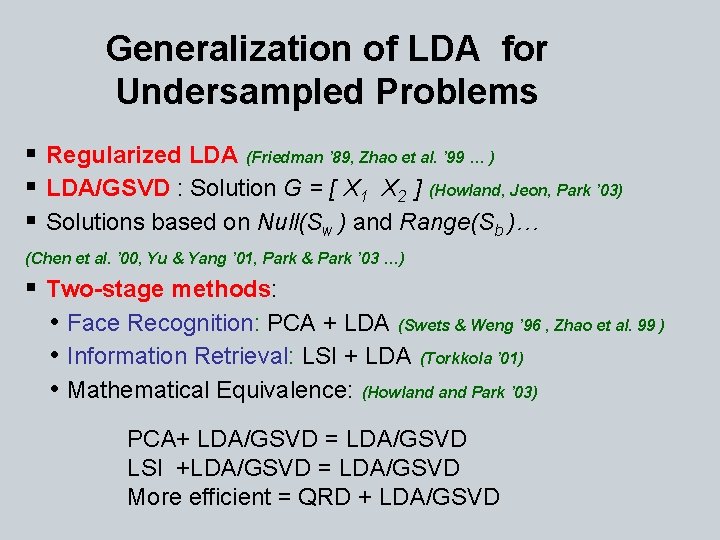
Generalization of LDA for Undersampled Problems § Regularized LDA (Friedman ’ 89, Zhao et al. ’ 99 … ) § LDA/GSVD : Solution G = [ X 1 X 2 ] (Howland, Jeon, Park ’ 03) § Solutions based on Null(Sw ) and Range(Sb )… (Chen et al. ’ 00, Yu & Yang ’ 01, Park & Park ’ 03 …) § Two-stage methods: • Face Recognition: PCA + LDA (Swets & Weng ’ 96 , Zhao et al. 99 ) • Information Retrieval: LSI + LDA (Torkkola ’ 01) • Mathematical Equivalence: (Howland Park ’ 03) PCA+ LDA/GSVD = LDA/GSVD LSI +LDA/GSVD = LDA/GSVD More efficient = QRD + LDA/GSVD

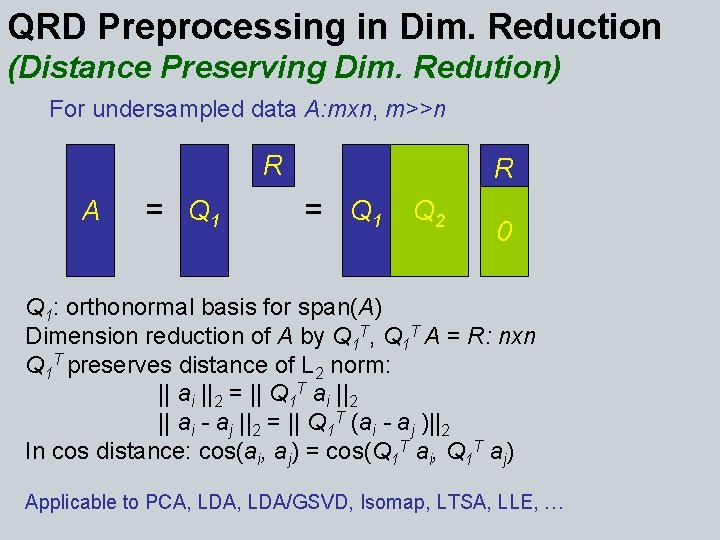
QRD Preprocessing in Dim. Reduction (Distance Preserving Dim. Redution) For undersampled data A: mxn, m>>n R A = Q 1 R = Q 1 Q 2 0 Q 1: orthonormal basis for span(A) Dimension reduction of A by Q 1 T, Q 1 T A = R: nxn Q 1 T preserves distance of L 2 norm: || ai ||2 = || Q 1 T ai ||2 || ai - aj ||2 = || Q 1 T (ai - aj )||2 In cos distance: cos(ai, aj) = cos(Q 1 T ai, Q 1 T aj) Applicable to PCA, LDA/GSVD, Isomap, LTSA, LLE, …

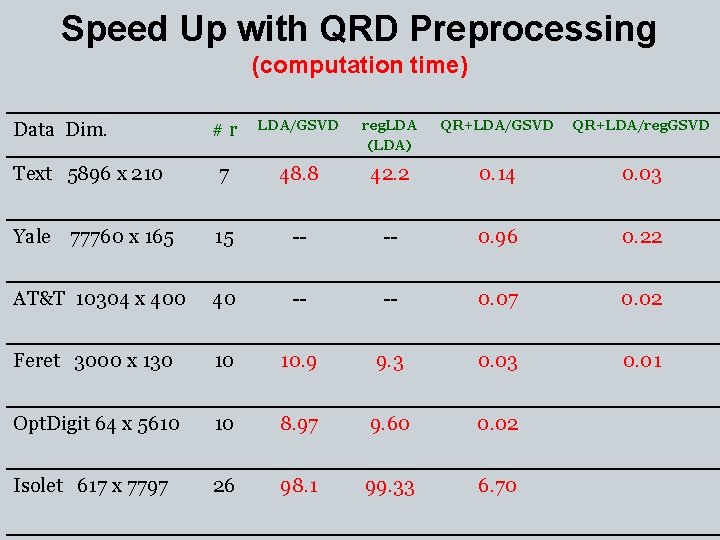
Speed Up with QRD Preprocessing (computation time) #r LDA/GSVD reg. LDA (LDA) QR+LDA/GSVD QR+LDA/reg. GSVD Text 5896 x 210 7 48. 8 42. 2 0. 14 0. 03 Yale 77760 x 165 15 -- -- 0. 96 0. 22 AT&T 10304 x 400 40 -- -- 0. 07 0. 02 Feret 3000 x 130 10 10. 9 9. 3 0. 01 Opt. Digit 64 x 5610 10 8. 97 9. 60 0. 02 Isolet 617 x 7797 26 98. 1 99. 33 6. 70 Data Dim.

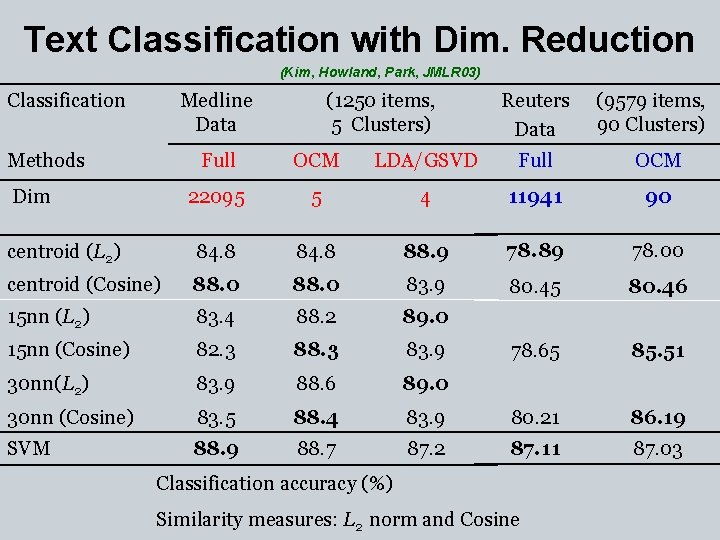
Text Classification with Dim. Reduction (Kim, Howland, Park, JMLR 03) Classification Medline Data (1250 items, 5 Clusters) Reuters Data (9579 items, 90 Clusters) Full OCM LDA/GSVD Full OCM 22095 5 4 11941 90 centroid (L 2) 84. 8 88. 9 78. 89 78. 00 centroid (Cosine) 88. 0 83. 9 80. 45 80. 46 15 nn (L 2) 83. 4 88. 2 89. 0 15 nn (Cosine) 82. 3 88. 3 83. 9 78. 65 85. 51 30 nn(L 2) 83. 9 88. 6 89. 0 30 nn (Cosine) 83. 5 88. 4 83. 9 80. 21 86. 19 SVM 88. 9 88. 7 87. 2 87. 11 87. 03 Methods Dim Classification accuracy (%) Similarity measures: L 2 norm and Cosine

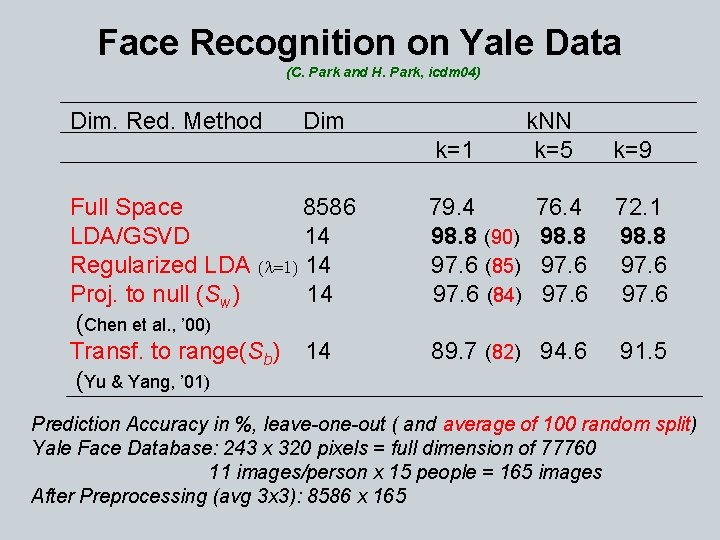
Face Recognition on Yale Data (C. Park and H. Park, icdm 04) Dim. Red. Method Dim k=1 Full Space 8586 LDA/GSVD 14 Regularized LDA (l=1) 14 Proj. to null (Sw) 14 (Chen et al. , ’ 00) Transf. to range(Sb) 14 (Yu & Yang, ’ 01) 79. 4 98. 8 (90) 97. 6 (85) 97. 6 (84) k. NN k=5 k=9 76. 4 98. 8 97. 6 72. 1 98. 8 97. 6 89. 7 (82) 94. 6 91. 5 Prediction Accuracy in %, leave-one-out ( and average of 100 random split) Yale Face Database: 243 x 320 pixels = full dimension of 77760 11 images/person x 15 people = 165 images After Preprocessing (avg 3 x 3): 8586 x 165

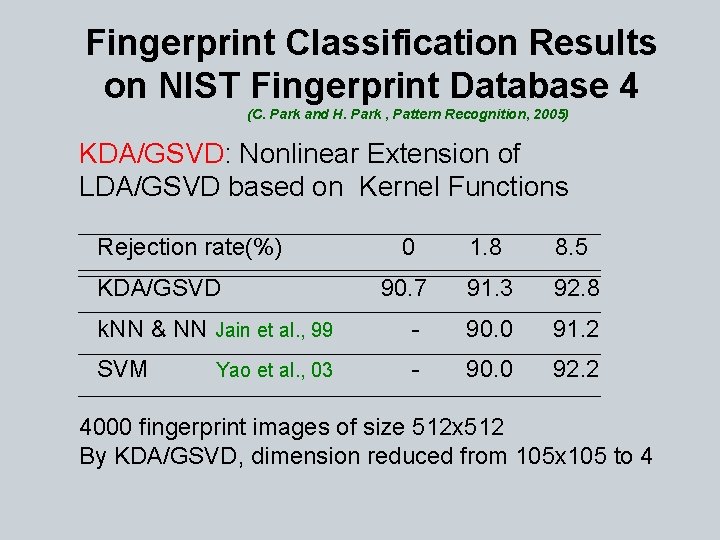
Fingerprint Classification Results on NIST Fingerprint Database 4 (C. Park and H. Park , Pattern Recognition, 2005) KDA/GSVD: Nonlinear Extension of LDA/GSVD based on Kernel Functions Rejection rate(%) KDA/GSVD 0 1. 8 8. 5 90. 7 91. 3 92. 8 k. NN & NN Jain et al. , 99 - 90. 0 91. 2 SVM - 90. 0 92. 2 Yao et al. , 03 4000 fingerprint images of size 512 x 512 By KDA/GSVD, dimension reduced from 105 x 105 to 4

Nonnegativity Preserving Dim. Reduction Nonnegative Matrix Factorization (Paatero&Tappa 94, Lee&Seung NATURE 99, Pauca et al. SIAM DM 04, Hoyer 04, Lin 05, Berry 06, Kim and Park 06, …) Given A: mxn with A>=0 and k << min (m, n), find W: mxk and H: kxn with W>=0 and H>=0 s. t. A ~ = H W min || A – WH ||F NMF/ANLS: Two-block Coordinate Descent Method in Bound-constrained Opt. Iterate the following ANLS (Kim and Park, Bioinformatics, to appear ) : fixing W , solve min. H>=0 || W H –A||F fixing H , solve min. W>=0 || HT WT –AT||F Any limit point is a stationary point (Grippo and Siandrone 00)

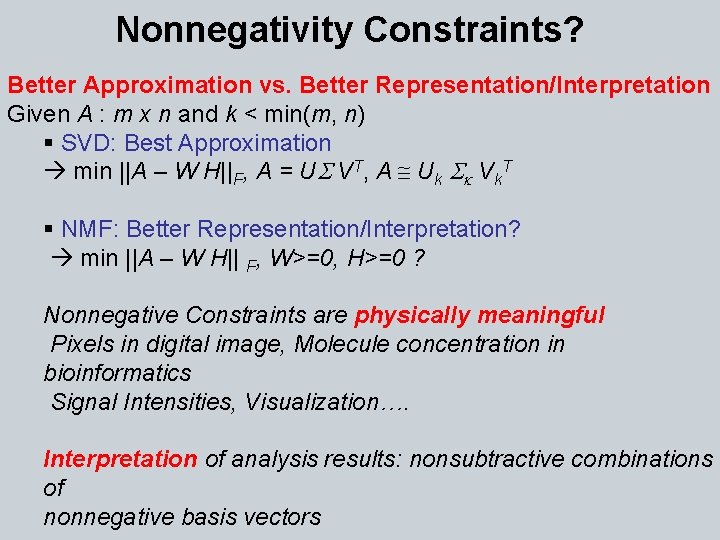
Nonnegativity Constraints? Better Approximation vs. Better Representation/Interpretation Given A : m x n and k < min(m, n) § SVD: Best Approximation min ||A – W H||F, A = US VT, A @ Uk Sk Vk. T § NMF: Better Representation/Interpretation? min ||A – W H|| F, W>=0, H>=0 ? Nonnegative Constraints are physically meaningful Pixels in digital image, Molecule concentration in bioinformatics Signal Intensities, Visualization…. Interpretation of analysis results: nonsubtractive combinations of nonnegative basis vectors

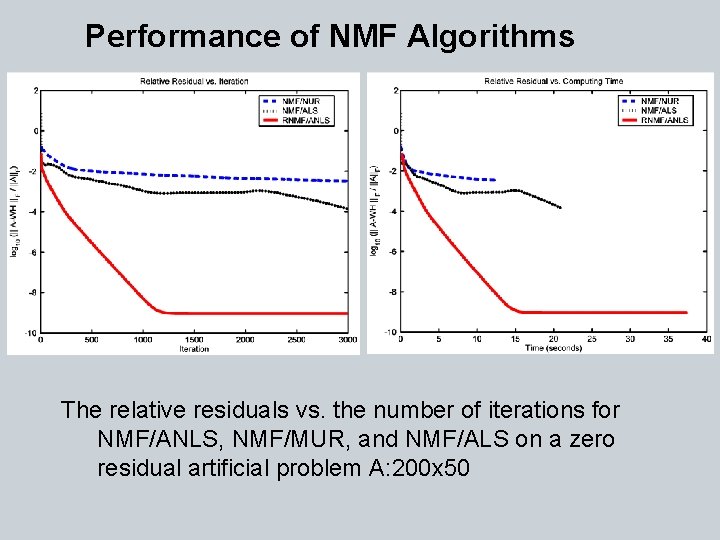
Performance of NMF Algorithms The relative residuals vs. the number of iterations for NMF/ANLS, NMF/MUR, and NMF/ALS on a zero residual artificial problem A: 200 x 50

Recovery of Factors by SVD and NMF A: 2500 x 28, W: 2500 x 3, H: 3 x 28 where A=W*H Recovery of the factors W and H by SVD and NMF/ANLS

Summary Effective Algorithms for Dimension Reduction and Matrix Decompositions that exploits prior knowledge • Design of New Algorithms: e. g. for undersampled data • Take Advantage of Prior Knowledge for Physically More Meaningful Modeling • Storage and Efficiency Issues for Massive Scale Data • Adaptive Algorithms * Applicable to a wide range of problems (Text classification, Facial recognition, Fingerprint classification, Gene class discovery in Microarray data, Protein secondary structure prediction … ) Thank you!