EBI Proteomics Services Team Standards Data and Tools

EBI Proteomics Services Team – Standards, Data, and Tools for Proteomics • • Henning Hermjakob European Bioinformatics Institute SME forum 2009 Vienna

Context Reactome Pathways En. Vision Annotation Integration and dissemination Uni. Proteins Archive Int. Act Interactions PRIDE Mass Spec DAS

The Proteomics Identifications Database (PRIDE) • Centralized, standards compliant, public data repository for proteomics identifications • Open source • Open data • 50, 287, 408 spectra • 2, 555, 194 protein identifications • Detailed annotation of meta-data • Jones, P, et al: PRIDE: new developments and new datasets. Nucleic Acids Res. 2008 Jan; 36(Database issue): D 878 -83. http: //www. ebi. ac. uk/pride
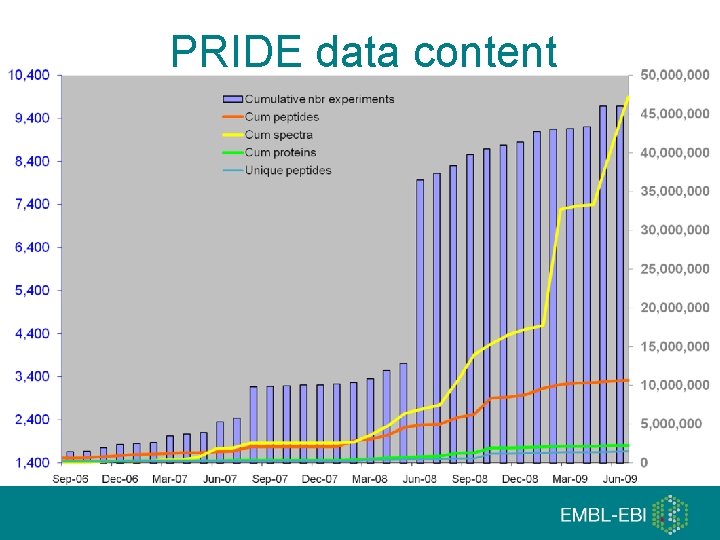
PRIDE data content
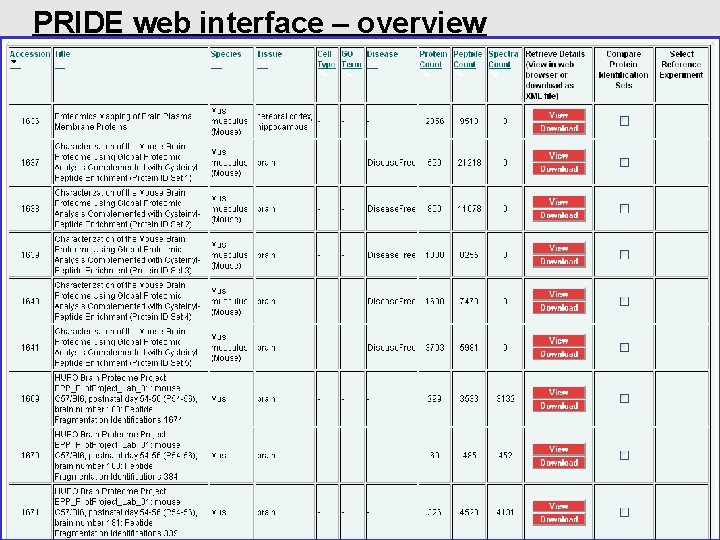
PRIDE web interface – overview pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride
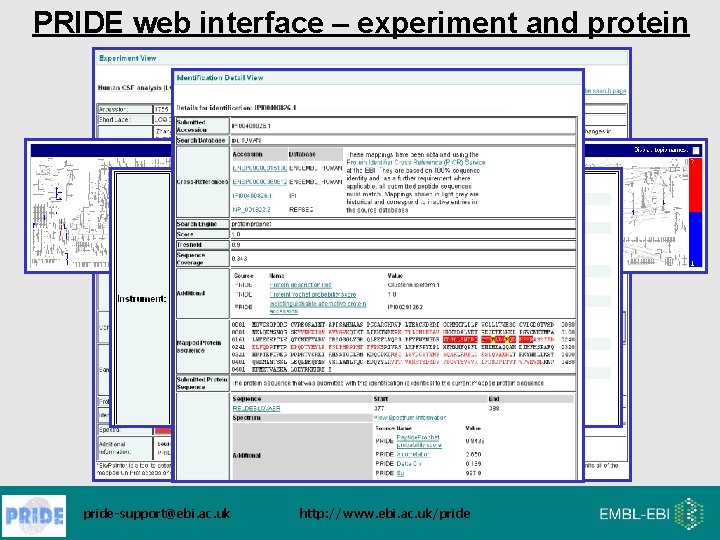
PRIDE web interface – experiment and protein pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride
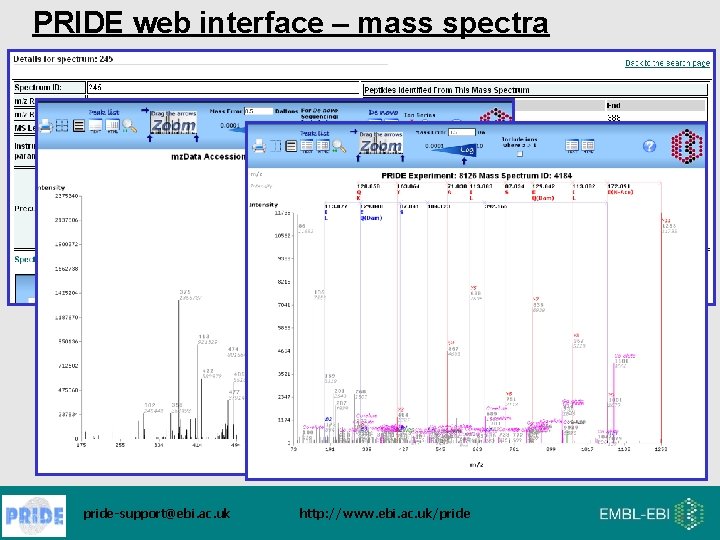
PRIDE web interface – mass spectra pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride
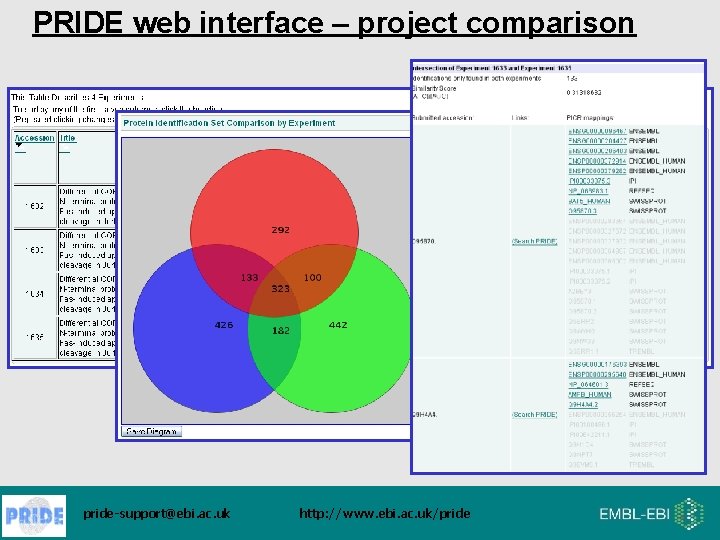
PRIDE web interface – project comparison pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride
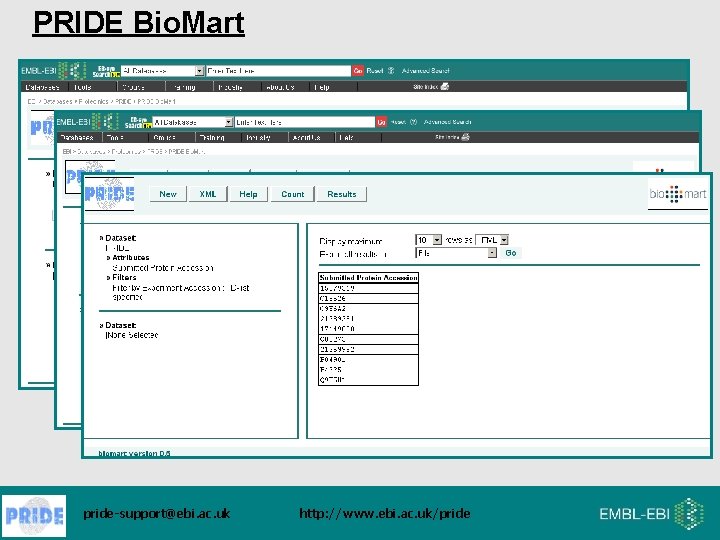
PRIDE Bio. Mart pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride
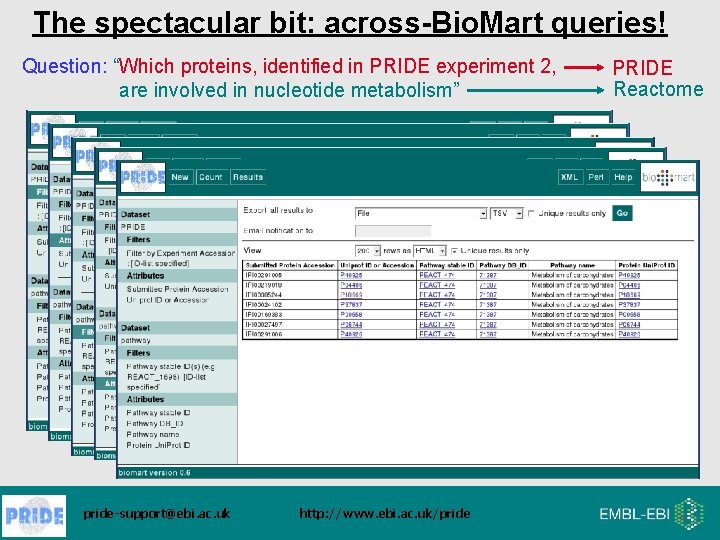
The spectacular bit: across-Bio. Mart queries! Question: “Which proteins, identified in PRIDE experiment 2, are involved in nucleotide metabolism” pride-support@ebi. ac. uk http: //www. ebi. ac. uk/pride PRIDE Reactome
The Int. Act Molecular Interaction Database • Centralized, standards compliant, public data repository for protein interactions • Open source • Open data • 200. 000 binary interaction reports • S. Kerrien, et al: Int. Act – Open Source Resource for Molecular Interaction Data. Nucleic Acids Res. 2007 Jan; 35(Database issue): D 561 -5. • Orchard, S. et al: The minimum information required for reporting a molecular interaction experiment (MIMIx). Nat Biotechnol. 2007 Aug; 25(8): 894 -8. http: //www. ebi. ac. uk/intact
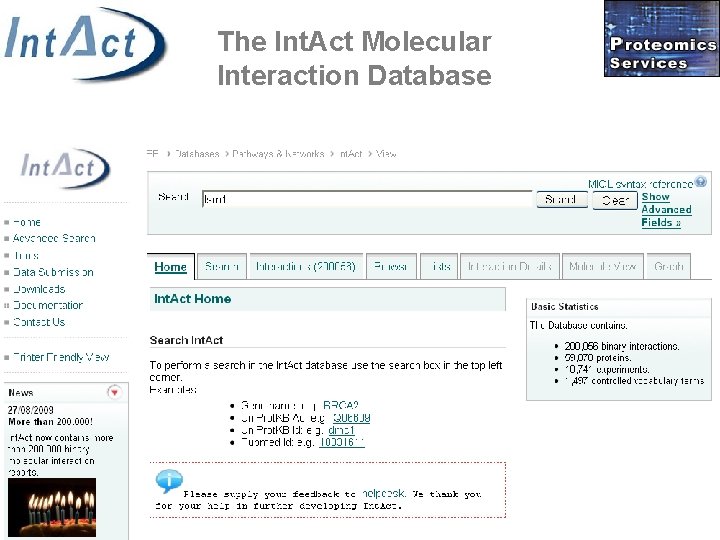
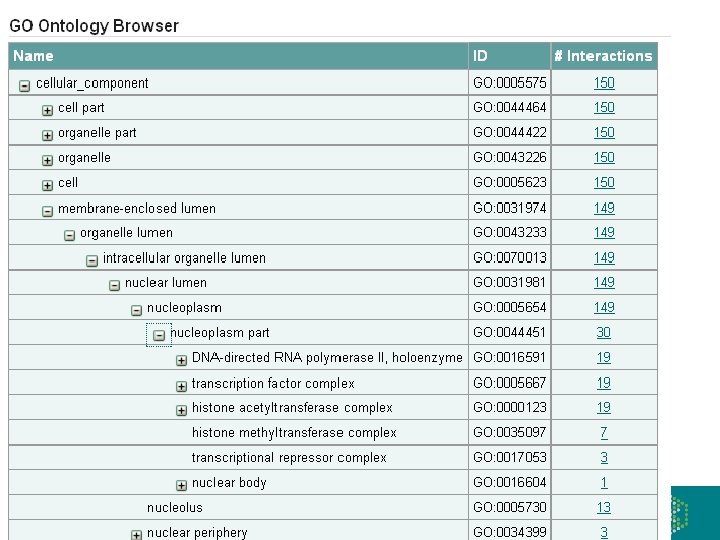
The Int. Act Molecular Interaction Database
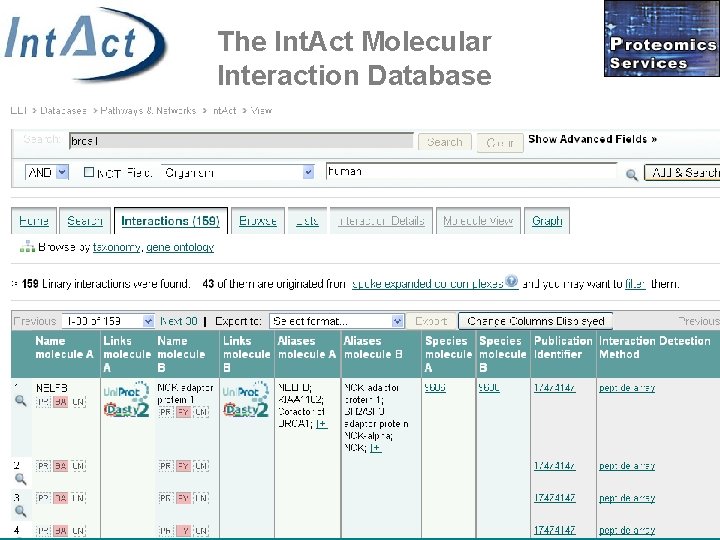
The Int. Act Molecular Interaction Database
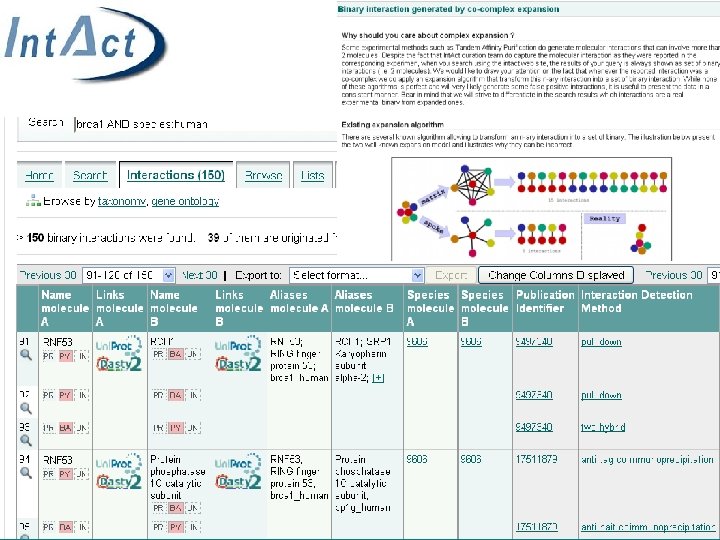
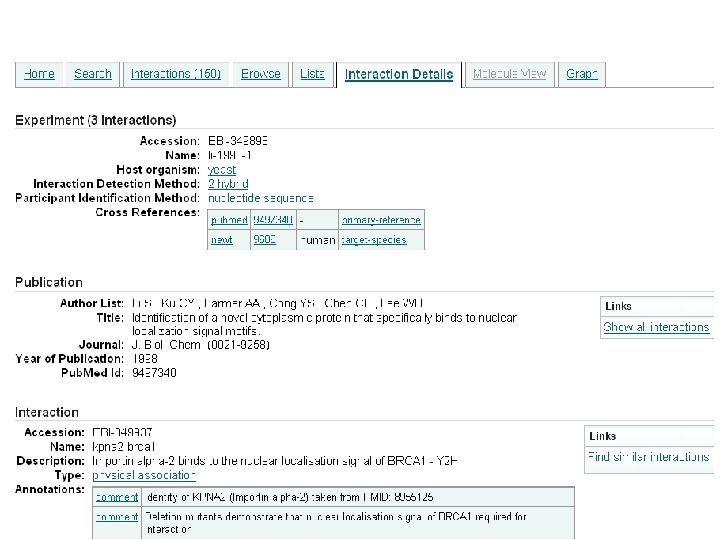

Reactome • Human pathway knowledgebase • Manually curated • Open source, open data • Collaboration between EBI, OCRI and NYU • Online since 2003 • Matthews L, et al: Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2008 Nov 3. http: //www. ebi. ac. uk/pride
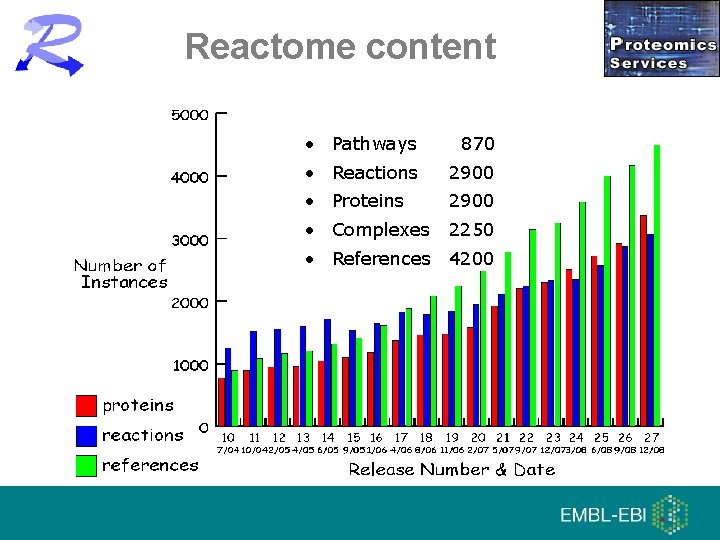
Reactome content • Pathways 870 • Reactions 2900 • Proteins 2900 • Complexes 2250 • References 4200
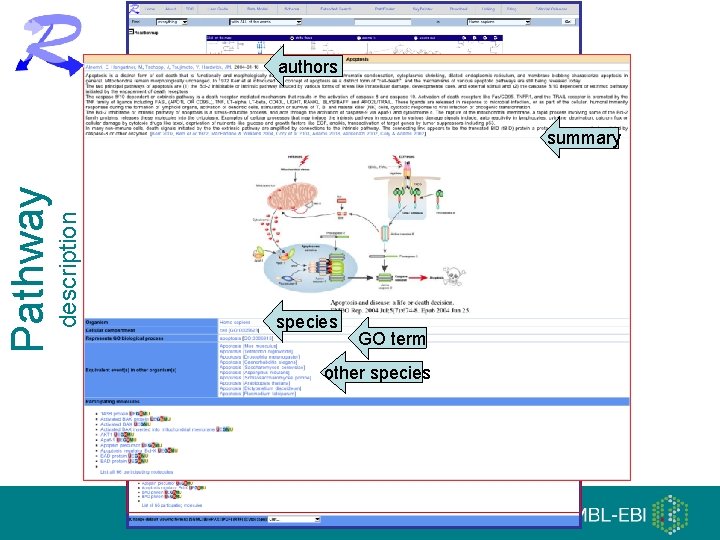
authors description Pathway summary species GO term other species
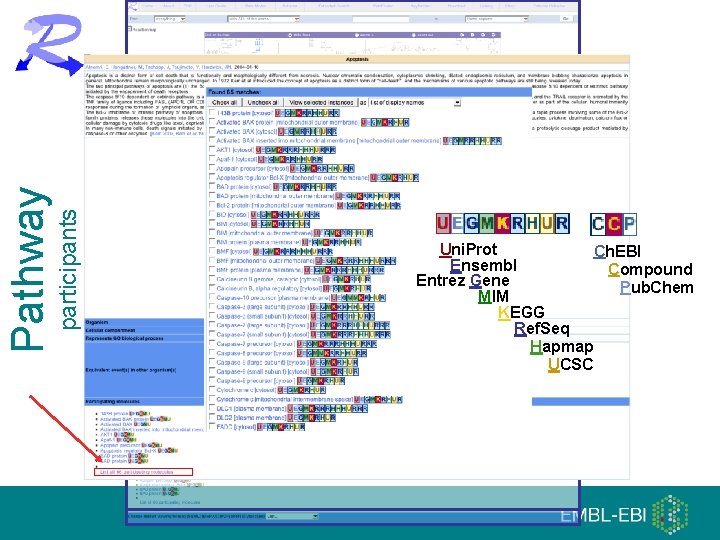
participants Pathway Uni. Prot Ch. EBI Ensembl Compound Entrez Gene Pub. Chem MIM KEGG Ref. Seq Hapmap UCSC
Sky. Painter • ‘Painting’ the reaction map with usersupplied data, e. g. over- and underexpressed genes from a microarray analysis • Animation for time series experiments • Overrepresentation analysis, e. g. disease candidate genes concentrated to a pathway
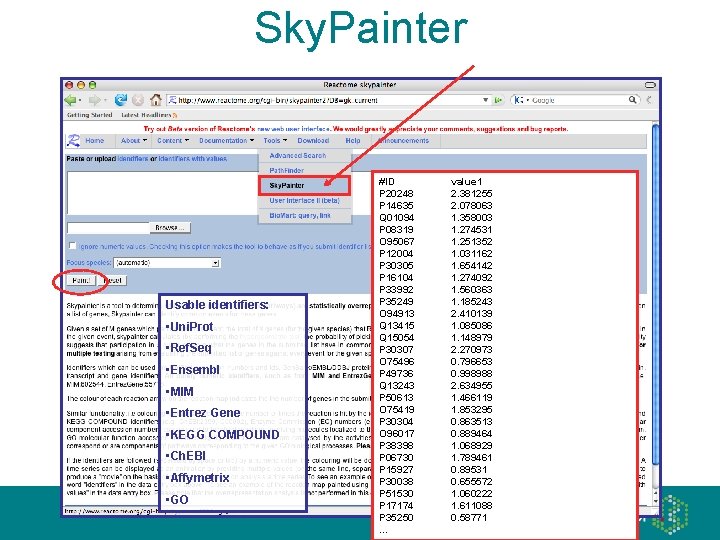
Sky. Painter Usable identifiers: • Uni. Prot • Ref. Seq • Ensembl • MIM • Entrez Gene • KEGG COMPOUND • Ch. EBI • Affymetrix • GO #ID P 20248 P 14635 Q 01094 P 08319 O 95067 P 12004 P 30305 P 16104 P 33992 P 35249 O 94913 Q 13415 Q 15054 P 30307 O 75496 P 49736 Q 13243 P 50613 O 75419 P 30304 O 96017 P 38398 P 06730 P 15927 P 30038 P 51530 P 17174 P 35250 … value 1 2. 381255 2. 078063 1. 358003 1. 274531 1. 251352 1. 031162 1. 654142 1. 274092 1. 560363 1. 185243 2. 410139 1. 085086 1. 148979 2. 270973 0. 796653 0. 998988 2. 634955 1. 466119 1. 853295 0. 863513 0. 889464 1. 068929 1. 789461 0. 89531 0. 655572 1. 060222 1. 611088 0. 58771
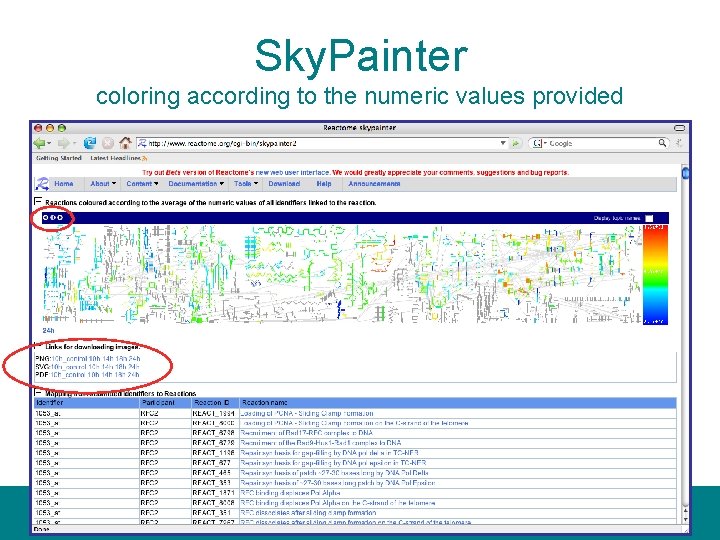
Sky. Painter coloring according to the numeric values provided
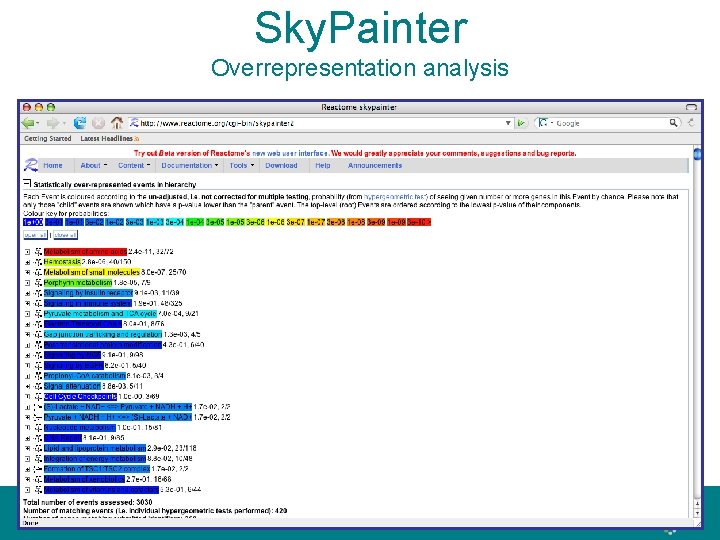
Sky. Painter Overrepresentation analysis
The Team The Funding • EU: – – – – Pro. Da. C (to 03/2009) Proteome. Binders Bio. Sapiens Felics Lipidomic. Net APO-SYS PSIMEx (since 03/2009) • EMBL • Wellcome Trust • NIH

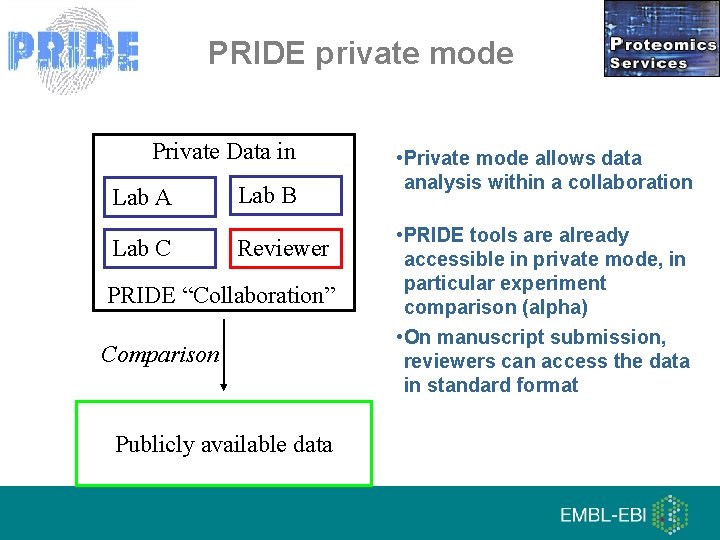
PRIDE private mode Private Data in Lab A Lab C Lab B Reviewer PRIDE “Collaboration” Comparison Publicly available data • Private mode allows data analysis within a collaboration • PRIDE tools are already accessible in private mode, in particular experiment comparison (alpha) • On manuscript submission, reviewers can access the data in standard format
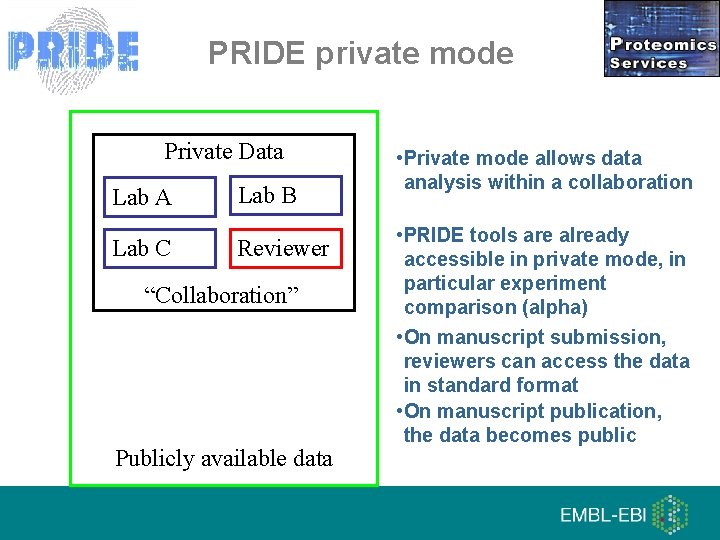
PRIDE private mode Private Data Lab A Lab C Lab B Reviewer “Collaboration” Publicly available data • Private mode allows data analysis within a collaboration • PRIDE tools are already accessible in private mode, in particular experiment comparison (alpha) • On manuscript submission, reviewers can access the data in standard format • On manuscript publication, the data becomes public
- Slides: 31