Drug Discovery Grid A real grid application Zhang




















- Slides: 43

Drug Discovery Grid -- A real grid application Zhang Wenju, Shen Jianhua Shanghai Institute of Materia Medica, CAS Shanghai Jiaotong University Jiangnan Institute of Computing The University of Hong Kong

Agenda 1. DDGrid Introduction 2. DDGrid Architecture 3. DDGrid Application 4. DDGrid Demo

Background Large-scale High-throughput Virtual Screening « in Silico The computational analysis of chemical databases to identify compounds appropriate for a given biological receptor « in Vitro Identification of new compounds showing some activity against a target biological receptor, and the progressive optimization of these leads to yield a compound with improved potency and physicochemical properties in vitro « in Vivo eventually, improved efficacy, pharmacokinetics, and toxicological profiles in vivo.

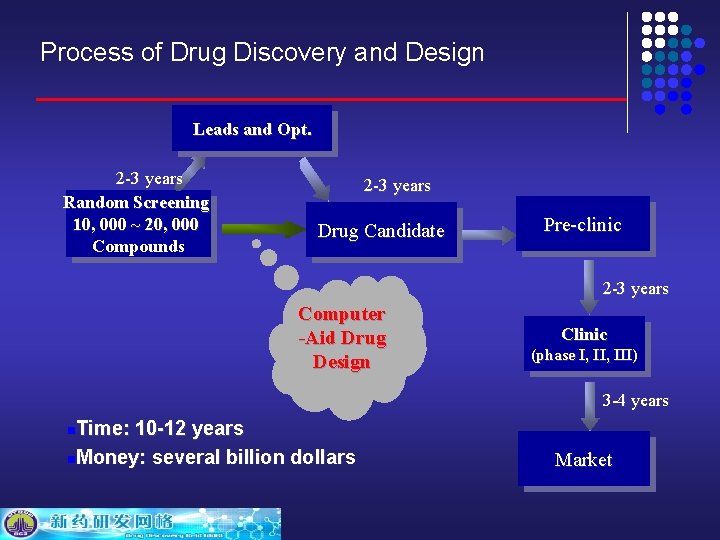
Process of Drug Discovery and Design Leads and Opt. 2 -3 years Random Screening 10, 000 ~ 20, 000 Compounds 2 -3 years Drug Candidate Pre-clinic 2 -3 years Computer -Aid Drug Design Clinic (phase I, III) 3 -4 years Time: 10 -12 years n. Money: several billion dollars n Market

DDGrid overview ◆ Drug Discovery Grid project aims to build a collaboration platform for drug discovery using the state-of-the-art grid computing technology. ◆ This project intends to solve large-scale computation and data intensive scientific applications in the fields of medicine chemistry and molecular biology with the help of grid middleware developed by our team. ◆ Over one million compounds database with 3 -D structure and physicochemical properties are also provided to identify potential drug candidates. Users also can build and maintain their own customized ligand database to share in this grid platform.

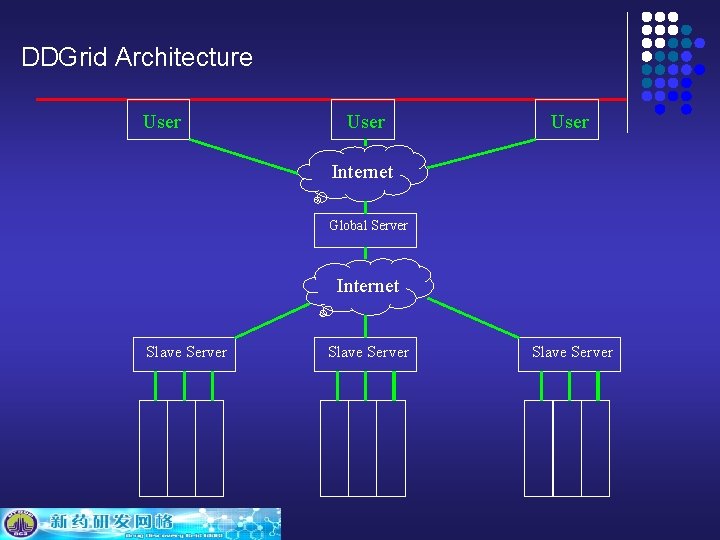
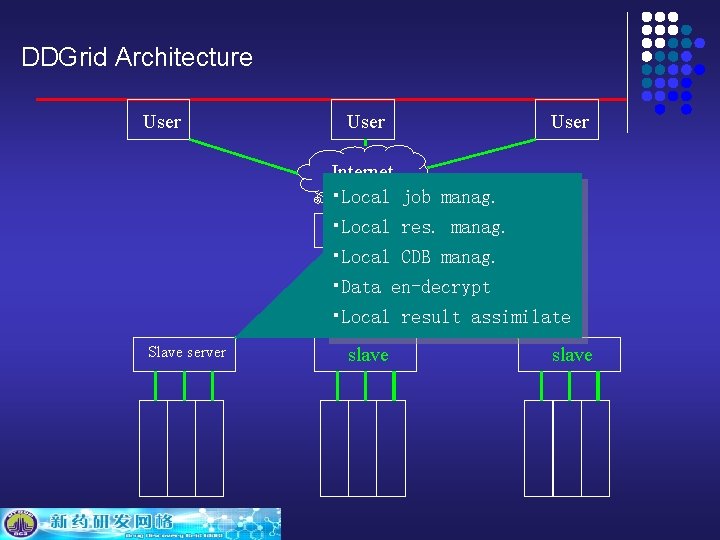
DDGrid Architecture User Internet Global Server Internet Slave Server

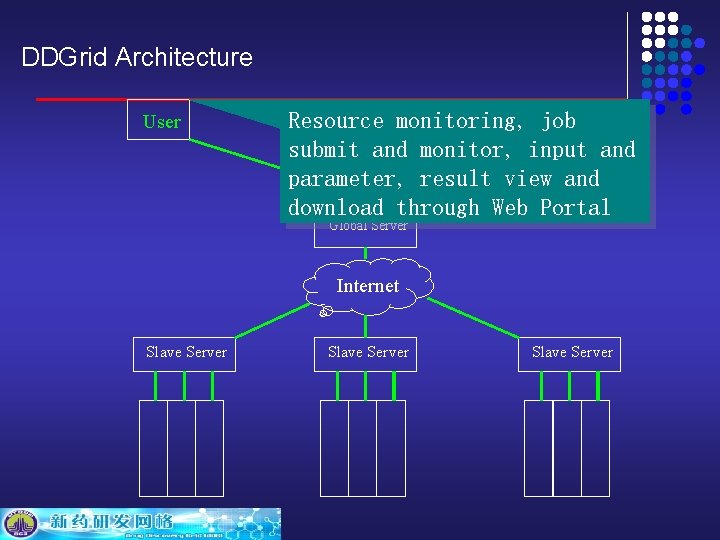
DDGrid Architecture User Resource 终端 monitoring, job 终端 submit and monitor, input and Internet result view and parameter, download through Web Portal Global Server Internet Slave Server

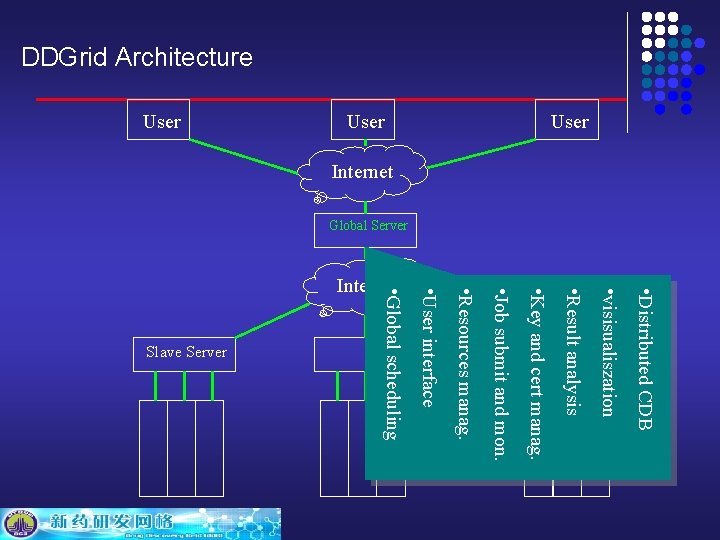
DDGrid Architecture • Distributed CDB • visisualiszation • User interface • Resources manag. • Job submit and mon. • Key and cert manag. • Result analysis 子服务器 Slave Server • Global scheduling Internet User Internet Global Server

DDGrid Architecture User Internet • Local job manag. • Local res. manag. 主服务器 • Local CDB manag. • Internet Data en-decrypt • Local result assimilate Slave server slave

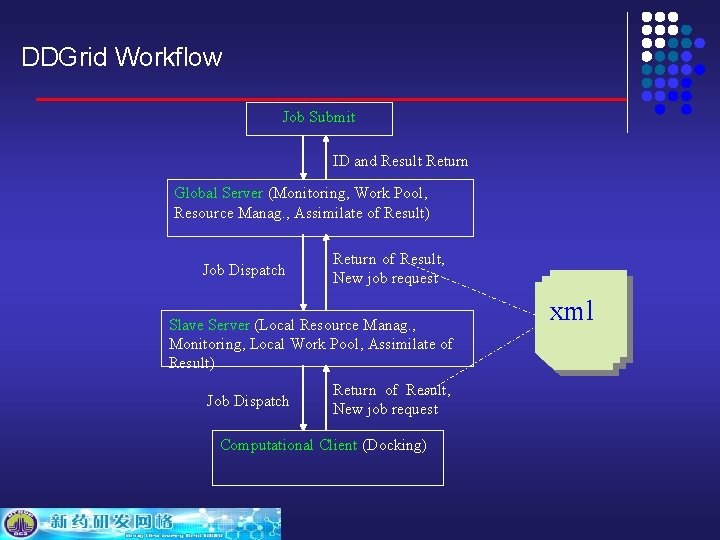
DDGrid Workflow Job Submit ID and Result Return Global Server (Monitoring, Work Pool, Resource Manag. , Assimilate of Result) Job Dispatch Return of Result, New job request Slave Server (Local Resource Manag. , Monitoring, Local Work Pool, Assimilate of Result) Job Dispatch Return of Result, New job request Computational Client (Docking) xml

DDGrid security 1. PKI-based security 2. All the sites involved should hold a certification issued by our CA 3. All the databases deployed and results are encrypted 4. All the message passing are SSL/TLS-enabled

DDGrid Web Portal

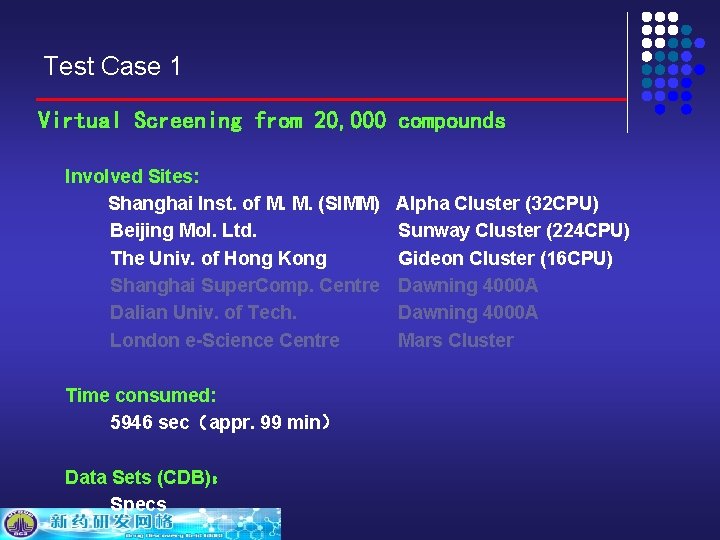
Test Case 1 Virtual Screening from 20, 000 compounds Involved Sites: Shanghai Inst. of M. M. (SIMM) Beijing Mol. Ltd. The Univ. of Hong Kong Shanghai Super. Comp. Centre Dalian Univ. of Tech. London e-Science Centre Time consumed: 5946 sec(appr. 99 min) Data Sets (CDB): Specs Alpha Cluster (32 CPU) Sunway Cluster (224 CPU) Gideon Cluster (16 CPU) Dawning 4000 A Mars Cluster

Job scheduling

Visualisation of Docking Result

DDGrid message passing <scheduler_request> <authenticator>3333</authenticator> <hostid>102</hostid> <rpc_seqno>2401</rpc_seqno> <platform_name>i 686 -pc-linux-gnu</platform_name> <core_client_major_version>2</core_client_major_version> <core_client_minor_version>19</core_client_minor_version> <idle_ncpu>16</idle_ncpu> <project_disk_usage>5315768. 000000</project_disk_usage> <total_disk_usage>68417940. 000000</total_disk_usage> <code_sign_key> … </code_sign_key> <projects> <project> <master_url>http: //www. ddgrid. ac. cn/ddg/</master_url> <resource_share>100. 000000</resource_share> </project> </projects> <result> … </result> … <host_info> … </host_info> </scheduler_request>

DDGrid message passing <scheduler_reply> <message priority="low">No work available</message> <project_name>Ddg</project_name> <user_name>sss</user_name> <code_sign_key> … </code_sign_key> … <workunit> … </workunit> <preferences> <low_water_days>1. 2</low_water_days> <high_water_days>2. 5</high_water_days> <disk_max_used_gb>0. 4</disk_max_used_gb> <disk_max_used_pct>50</disk_max_used_pct> <disk_min_free_gb>0. 4</disk_min_free_gb> … </preferences> … </scheduler_reply>

DDGrid message passing <workunit> <file_info> <number>0</number> </file_info> <file_info> <number>1</number> </file_info> <file_info> <number>2</number> </file_info> … <file_ref> <file_number>0</file_number> <open_name>tabfile</open_name> </file_ref> <file_ref> <file_number>1</file_number> <open_name>infile</open_name> </file_ref> <file_ref> <file_number>2</file_number> <open_name>sphfile</open_name> </file_ref> <command_line>-business</command_line> </workunit>

DDGrid message passing … <project> <scheduler_url>http: //www. ddgrid. ac. cn/ddg_cgi/cgi</scheduler_url> <master_url>http: //www. ddgrid. ac. cn/ddg/</master_url> <project_name>Ddg</project_name> </project> <app> <name>gridapp</name> </app> <file_info> <name>gridapp/gridapp_2. 19_i 686 -pc-linux-gnu</name> <nbytes>260754. 000000</nbytes> <max_nbytes>0. 000000</max_nbytes> <executable/> <signature_required/> <file_signature> … </file_signature> <url>http: //www. ddgrid. ac. cn/ddg/download/gridapp_2. 19_i 686 -pc-linux-gnu</url> </file_info> <file_info> … </file_info> …

DDGrid Resources Computational and Data Resources Integration Resources aggregated SIMM Sunway 32 A Cluster Beijing Molecule Inc. Sunway 256 P Cluster HKU Gideon 300 Cluster SSC Dawning 4000 A Le. SC Mars Cluster (Test only) Singapore Poly-tech Univ. Dalian Univ. of Technology Shanghai Jiaotong Univ. Heterogeneous resources OS: IRIX, Digital Unix, Linux(IA 32, x 86_64) CPU:R 12000, Alpha, Pentium, AMD

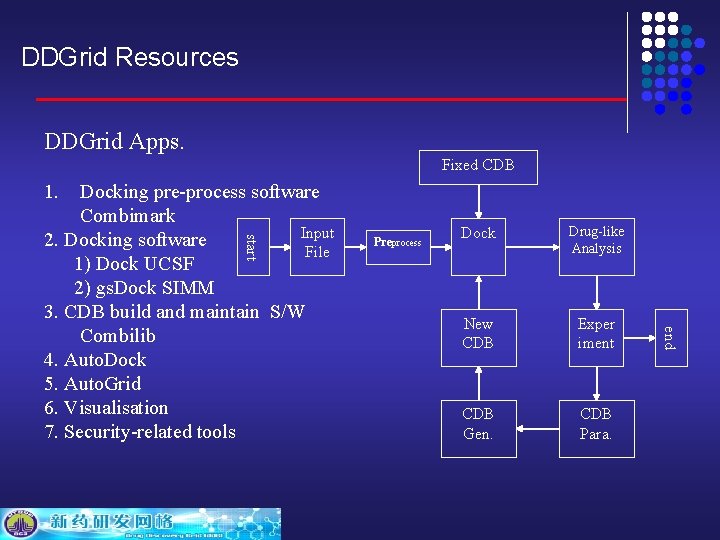
DDGrid Resources DDGrid Apps. Fixed CDB 1. start Preprocess Dock Drug-like Analysis New CDB Exper iment CDB Gen. CDB Para. end Docking pre-process software Combimark Input 2. Docking software File 1) Dock UCSF 2) gs. Dock SIMM 3. CDB build and maintain S/W Combilib 4. Auto. Dock 5. Auto. Grid 6. Visualisation 7. Security-related tools

DDGrid Resources Chemical Databases (CDB) Each ligand record in a chemical database represents the 3 D structural information of a compound. The numbers of compounds in each CDB can be in the order of tens of thousands and the database size be anywhere from tens of megabytes to gigabytes and even terabytes. 1. static databases purchased from commercial chemical company. Available Chemical Directory (ACD) Chinese natural product database (CNPD) SPECS database chemical ADME/T database, etc. 2. dynamic databases made by user own, and deployed automatically.

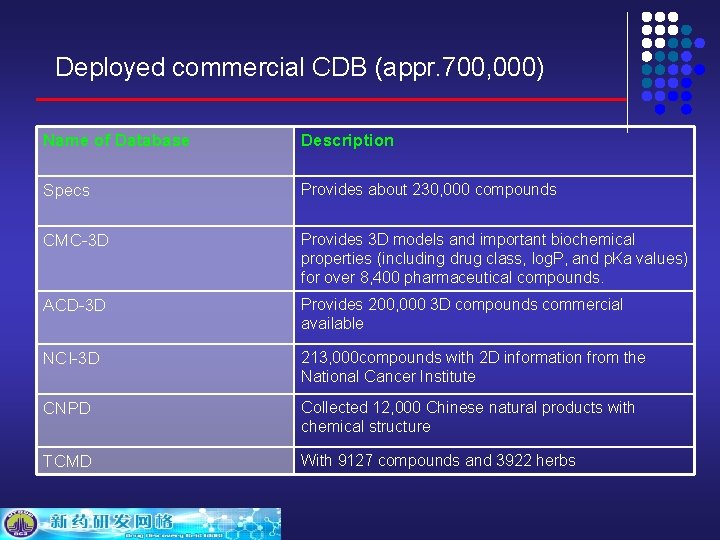
Deployed commercial CDB (appr. 700, 000) Name of Database Description Specs Provides about 230, 000 compounds CMC-3 D Provides 3 D models and important biochemical properties (including drug class, log. P, and p. Ka values) for over 8, 400 pharmaceutical compounds. ACD-3 D Provides 200, 000 3 D compounds commercial available NCI-3 D 213, 000 compounds with 2 D information from the National Cancer Institute CNPD Collected 12, 000 Chinese natural products with chemical structure TCMD With 9127 compounds and 3922 herbs

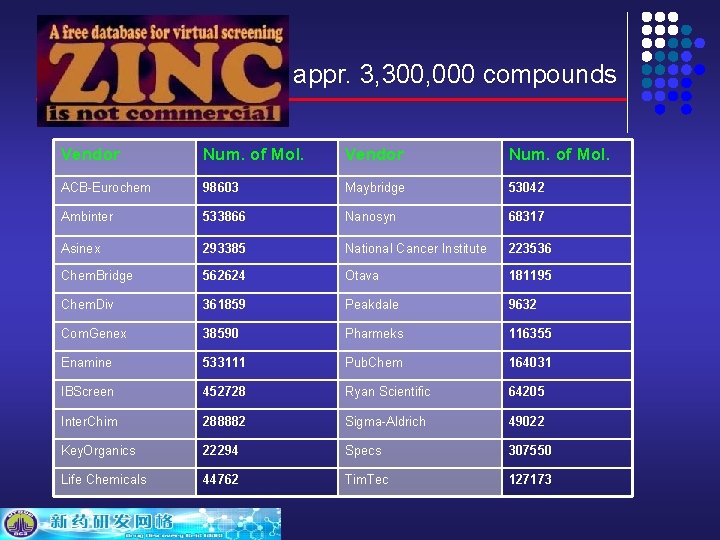
appr. 3, 300, 000 compounds Vendor Num. of Mol. ACB-Eurochem 98603 Maybridge 53042 Ambinter 533866 Nanosyn 68317 Asinex 293385 National Cancer Institute 223536 Chem. Bridge 562624 Otava 181195 Chem. Div 361859 Peakdale 9632 Com. Genex 38590 Pharmeks 116355 Enamine 533111 Pub. Chem 164031 IBScreen 452728 Ryan Scientific 64205 Inter. Chim 288882 Sigma-Aldrich 49022 Key. Organics 22294 Specs 307550 Life Chemicals 44762 Tim. Tec 127173

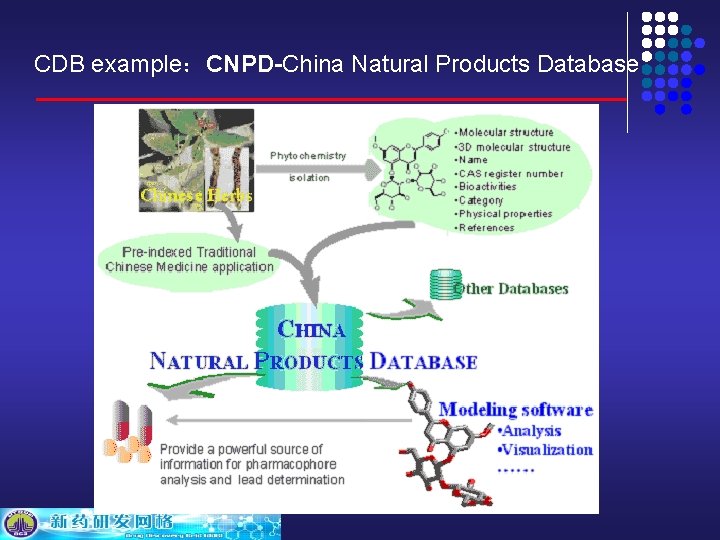
CDB example:CNPD-China Natural Products Database

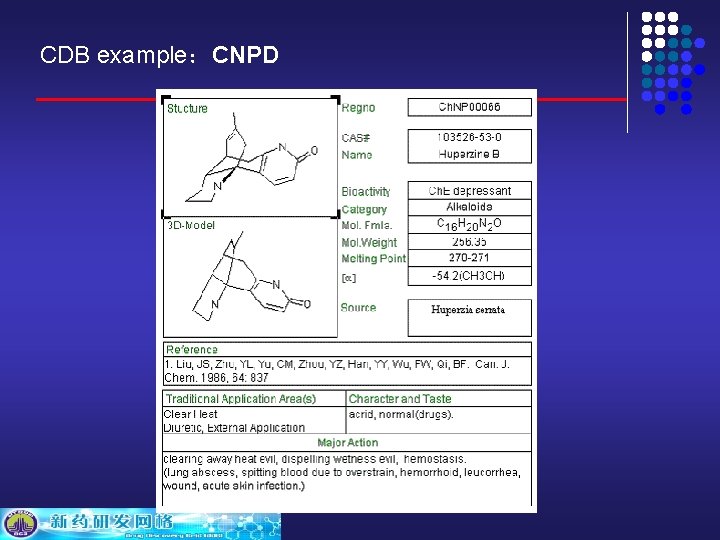
CDB example:CNPD: The first and only comprehensive source of chemical, structural and bibliographic data on all known natural products in China. CNPD serves as information sources for chemical, physical and biological properties, literature, they are useful to scientists within the pharmaceutical industry. CNPD can be searched in flexible ways: structure, sub-structure, name, molecular formula, molecular weight, CAS register number, category, etc. CNPD: Traditional Chinese Medicine (TCM) applications are preindexed in CNPD to provide hints for lead compounds discovery.

CDB example:CNPD

CDB example:TCMD-Traditional Chinese Medicine Database TCMD is a bibliographical database of approximately 20, 000 records with abstracts of TCM articles. Relevant articles are selected from among 150 -200 journals from Mainland China, Taiwan, and Hong Kong (most of them are Chinese); English abstracts are written for the selected articles and other pertinent information is translated into English.

CDB example:TCMD

DDGrid applications in reality SIMM carried out anti-SARS and anti-diabetes drug research using the DDGrid 1. Anti-SARS drug research 2. Anti-diabetes drug research

Research on Anti-SARS medicine Virtual screening from Comprehensive Medicinal Chemistry 3 D (CMC-3 D) database which contains 7, 900 compounds, found that cinanserin have distinct anti-SARS effect Department of Virology, Bernhard-Nocht-Institute for Tropical Medicine, Germany Research Department, Cantonal Hospital St Gallen, Switzerland “Basically your inhibitor turned out to be the best compound we have tested so far! ” Have applied for domestic patent 03129071. x and PCT patent pi 034248

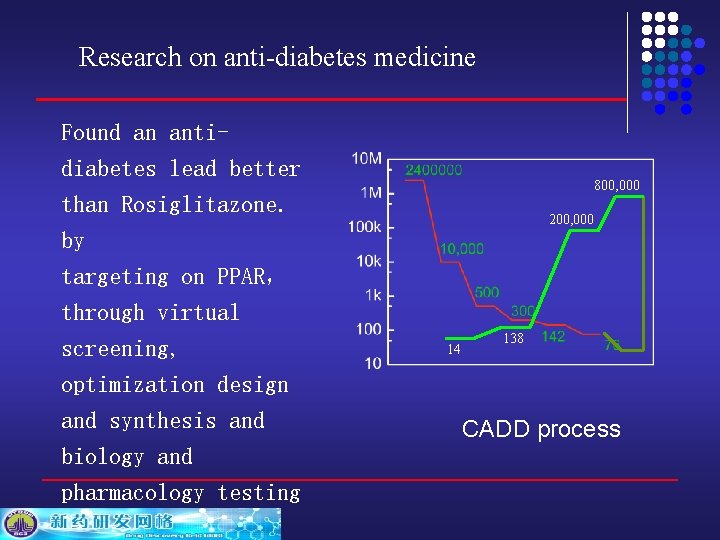
Research on anti-diabetes medicine Found an antidiabetes lead better than Rosiglitazone. by targeting on PPAR, through virtual screening, optimization design and synthesis and biology and pharmacology testing 800, 000 200, 000 14 138 CADD process

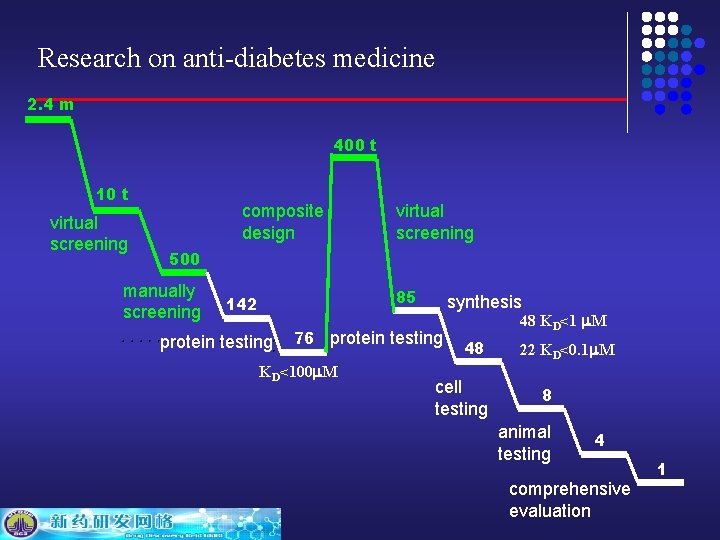
Research on anti-diabetes medicine 2. 4 m 400 t 10 t virtual screening composite design virtual screening 500 manually screening 85 142 protein testing synthesis 76 protein testing KD<100 m. M 48 KD<1 m. M 48 cell testing 22 KD<0. 1 m. M 8 animal testing 4 comprehensive evaluation 1

New anti-diabetes drug Current Progress 1. Applied for patent 200410016460. X,and PCT patent 2. Security testing and pre-clinic research

What does the DDGrid provide? 1、 Drug Design Collaboration Platform Large-scale Virtual Screening platform sharing large CDB 2、Computational Resources Sharing SIMM/SSC/HKU/Mol. Ltd/SJTU/DUT 3、Data Resources Sharing pre-deployed commercial CDB (ACD/CNPD …) sharing self-made CDB 4、Medicinal chemistry text and structure search 5、Customization and Extension

Collaboration Selected Users of DDGrid

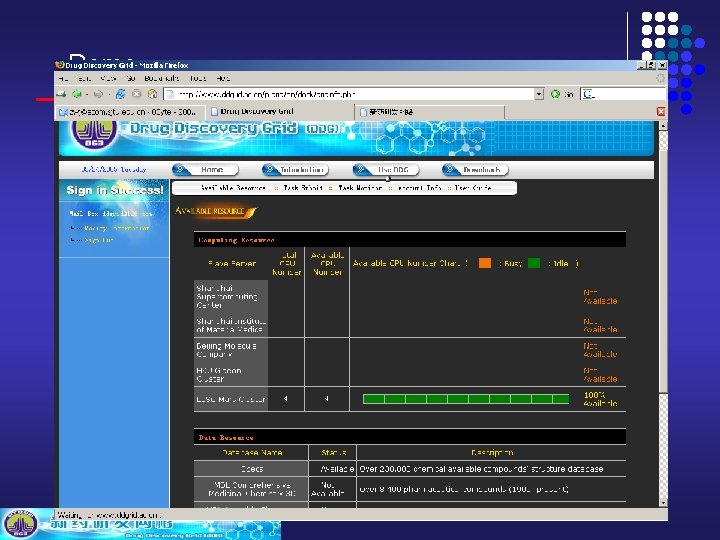
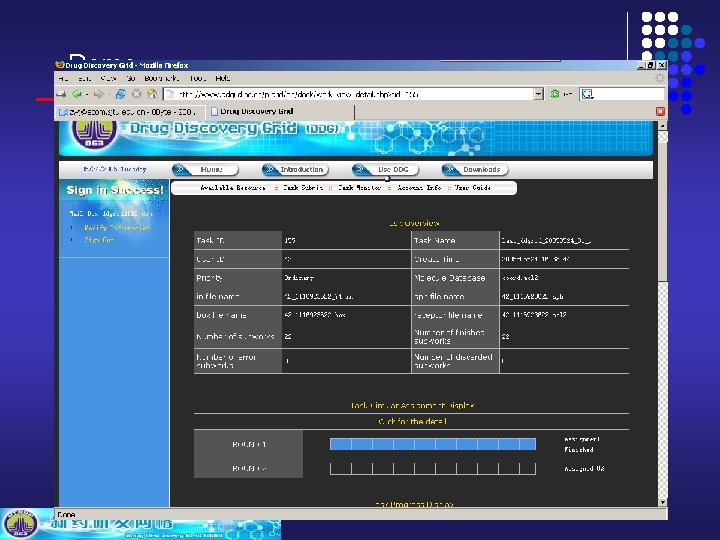
Demo DDGrid Demo http: //www. ddgrid. ac. cn

Demo

Demo

Demo

Demo

Demo

Q&A Thank you!