Drawing Darwins Diagram Creating Cladograms Carolus Linnaeus aka






- Slides: 23

Drawing Darwin’s Diagram: Creating Cladograms

Carolus Linnaeus (aka Carl von Linné) Swedish Botanist, Physician, and Zoologist 1707 - 1778 Developed a hierarchial classification scheme do deal with all these organisms, and we use to this day (though we are moving away from it a little bit)! http: //upload. wikimedia. org/wikipedia/commons/6/68/Carl_von_Linné. jpg

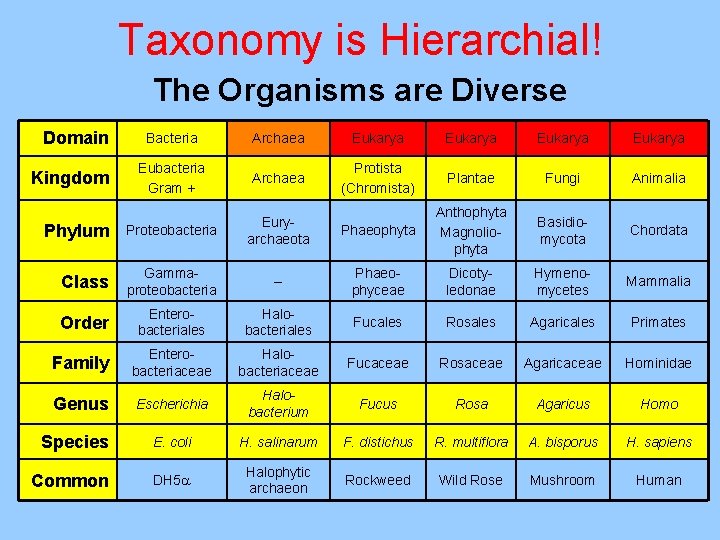
Taxonomy is Hierarchial! The Organisms are Diverse Domain Bacteria Archaea Eukarya Eubacteria Gram + Archaea Protista (Chromista) Plantae Fungi Animalia Phylum Proteobacteria Euryarchaeota Phaeophyta Anthophyta Magnoliophyta Basidiomycota Chordata Class Gammaproteobacteria -- Phaeophyceae Dicotyledonae Hymenomycetes Mammalia Order Enterobacteriales Halobacteriales Fucales Rosales Agaricales Primates Family Enterobacteriaceae Halobacteriaceae Fucaceae Rosaceae Agaricaceae Hominidae Genus Escherichia Halobacterium Fucus Rosa Agaricus Homo Species E. coli H. salinarum F. distichus R. multiflora A. bisporus H. sapiens Common DH 5 Halophytic archaeon Rockweed Wild Rose Mushroom Human Kingdom

Charles Darwin - British Naturalist Formal Studies: Medicine and Theology 1809 - 1882 Descent with Modification http: //www 2. biologie. unihalle. de/zool/mol_ecol/Bilder/Darwin. Bild. jpg 1880 The Power of Movement in Plants 1871 Descent of Man 1859 Origin of Species http: //upload. wikimedia. org/wikipedia/en/3/3 e/Charles_Darwin_1854. jpg Species evolved from generation to generation over time 1831 -1836 HMS Beagle Voyage http: //www. omniscopic. com/blog/uploaded_images/darwin-791061. jpg

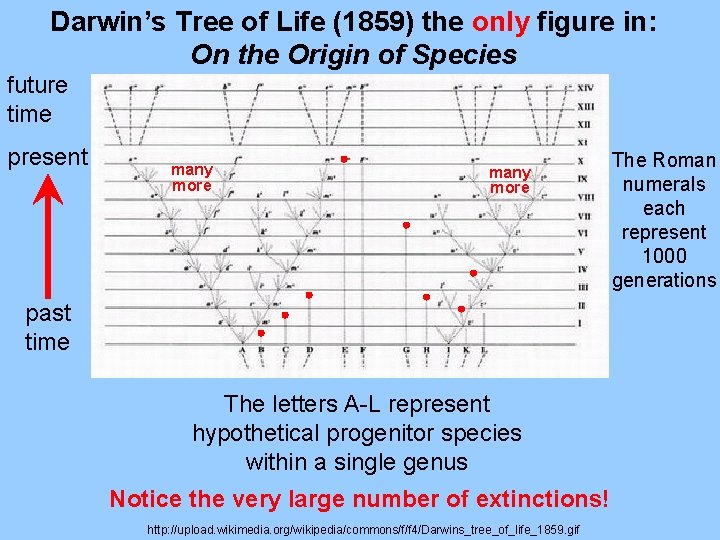
Darwin’s Tree of Life (1859) the only figure in: On the Origin of Species future time present many more past time The letters A-L represent hypothetical progenitor species within a single genus Notice the very large number of extinctions! http: //upload. wikimedia. org/wikipedia/commons/f/f 4/Darwins_tree_of_life_1859. gif The Roman numerals each represent 1000 generations

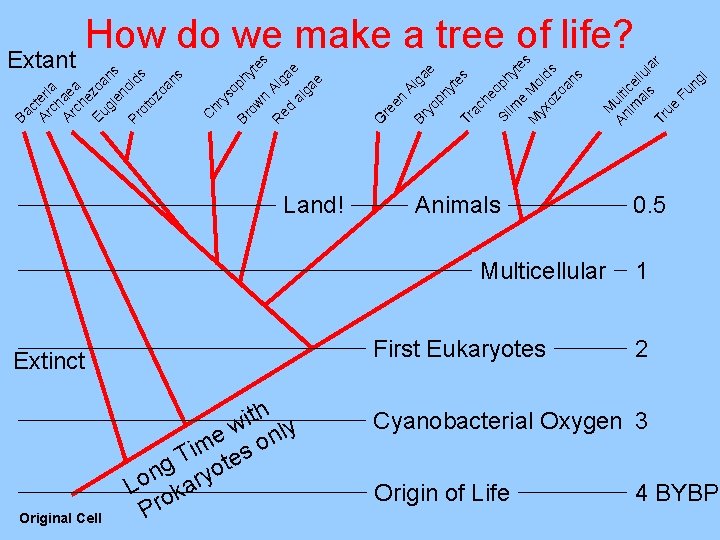
Original Cell Land! Extinct h t i w nly e im es o T g yot n Lo kar Pro es First Eukaryotes Origin of Life M An ulti im ce al llul s ar Tr ue Fu ng i he Sl oph im yt e e M Mo s yx oz lds oa ns ac Tr e ga Al ph yt yo Br en G re s te op hy A R ed lga e al ga e n ow Br ys C hr c Ar teri ch a Ar ae ch a e Eu zoa n gl en s oi Pr ds ot oz oa ns Ba How do we make a tree of life? Extant Animals Multicellular 0. 5 1 2 Cyanobacterial Oxygen 3 4 BYBP

Emil Hans (Willi) Hennig German entomologist 1913 -1976 Hennig developed a mechanism (cladistics) to find the pathways of evolution among related organisms. It is based not only on what one sees, but on many kinds of evidence, including molecular sequences. The pathways are determined by virtue of shared derived characteristics (synapomorphies). http: //scienceblogs. com/evolvingthoughts/upload/2007/04/Willi_Hennig 2. jpg Rather than putting organisms into Linnean taxonomic “boxes, ” the cladistics process shows the pathway of evolution.

Evidence Categories • History - clearer recently, more obscure anciently • Fossils - stratigraphic depth, isotope decay, etc. • Chemical - metabolic products such as O 2, Ss • Molecular - DNA sequence alterations, etc. • Developmental sequences - onto- phylo- geny • Biogeography - Pangea, Gondwana & Laurasia

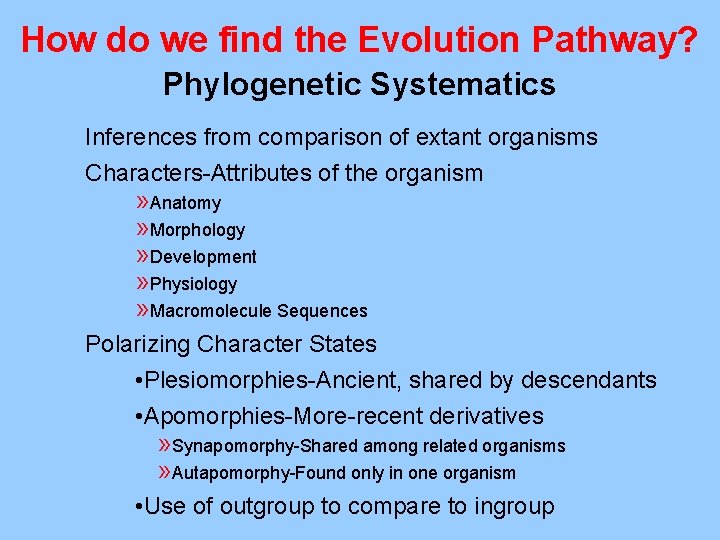
How do we find the Evolution Pathway? Phylogenetic Systematics Inferences from comparison of extant organisms Characters-Attributes of the organism » Anatomy » Morphology » Development » Physiology » Macromolecule Sequences Polarizing Character States • Plesiomorphies-Ancient, shared by descendants • Apomorphies-More-recent derivatives » Synapomorphy-Shared among related organisms » Autapomorphy-Found only in one organism • Use of outgroup to compare to ingroup

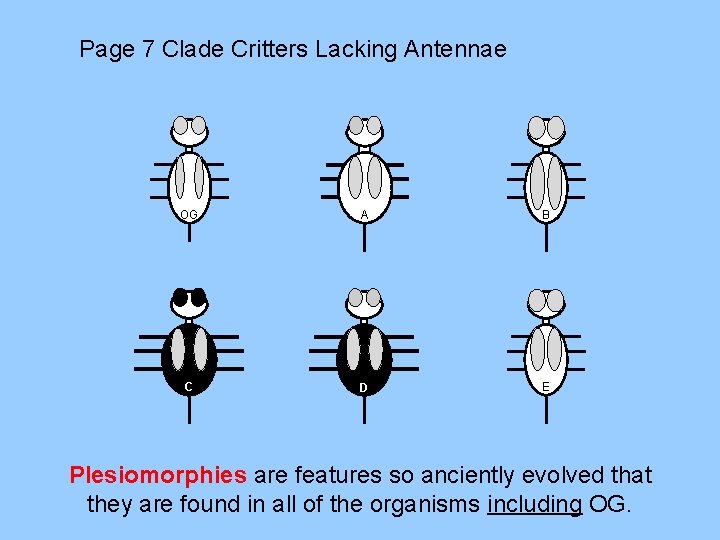
Page 7 Clade Critters Lacking Antennae OG A B C D E Plesiomorphies are features so anciently evolved that they are found in all of the organisms including OG.

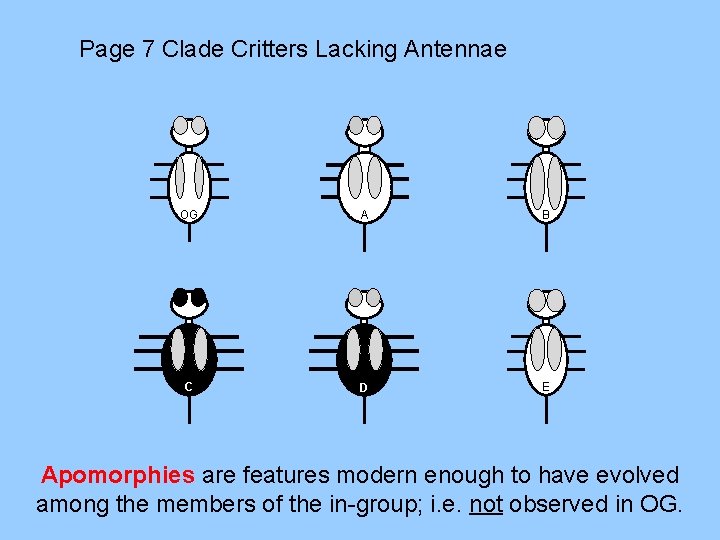
Page 7 Clade Critters Lacking Antennae OG A B C D E Apomorphies are features modern enough to have evolved among the members of the in-group; i. e. not observed in OG.

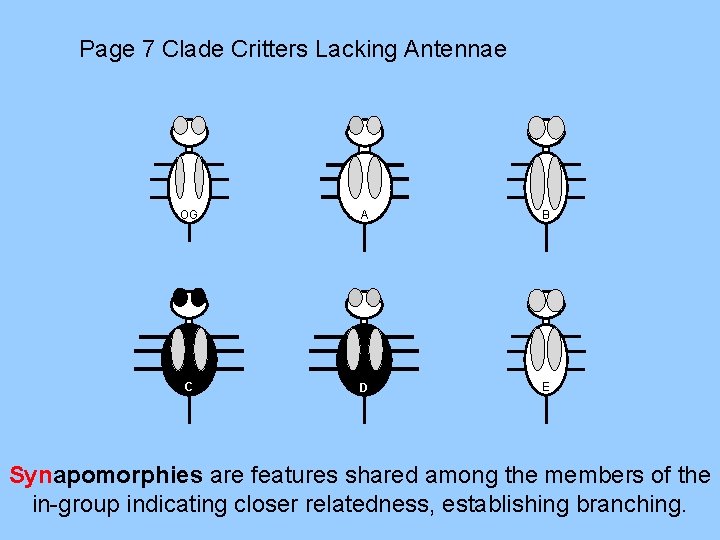
Page 7 Clade Critters Lacking Antennae OG A B C D E Synapomorphies are features shared among the members of the in-group indicating closer relatedness, establishing branching.

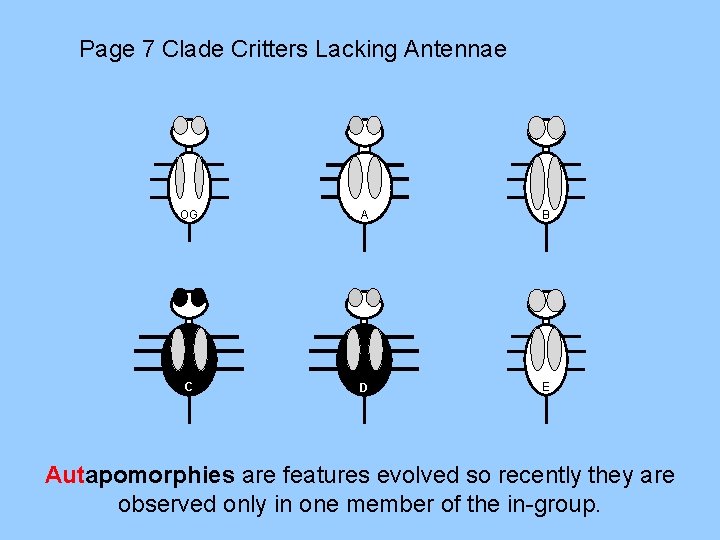
Page 7 Clade Critters Lacking Antennae OG A B C D E Autapomorphies are features evolved so recently they are observed only in one member of the in-group.

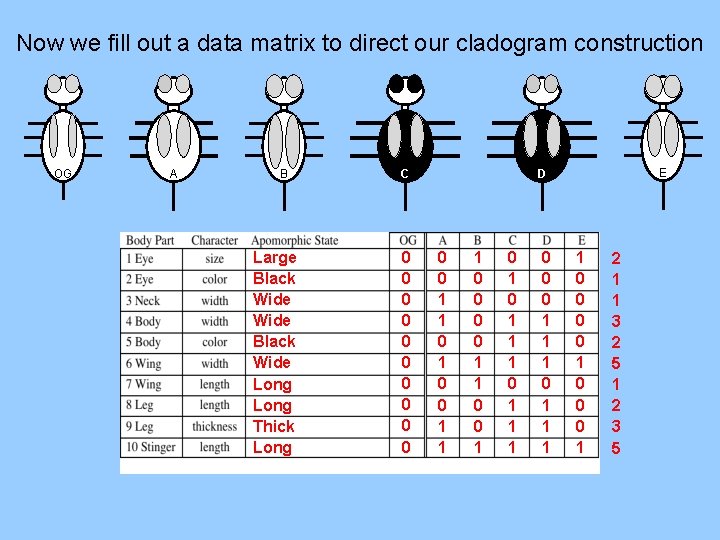
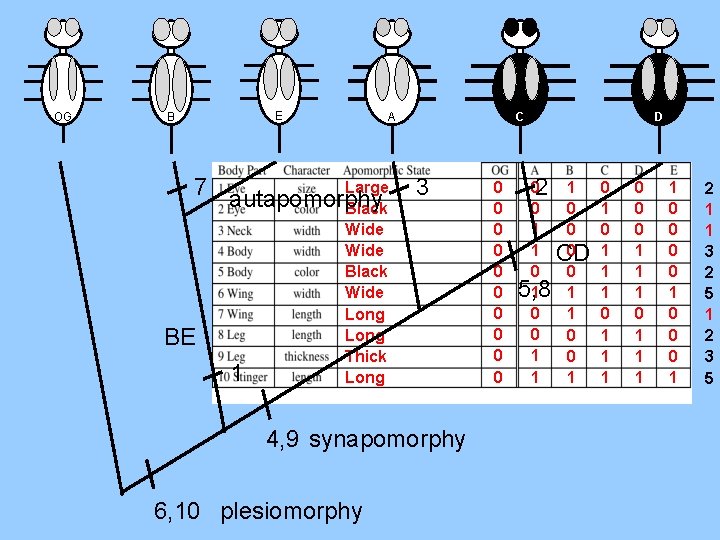
Now we fill out a data matrix to direct our cladogram construction OG A B Large Black Wide Long Thick Long C 0 0 0 0 0 E D 0 0 1 1 1 0 0 1 0 1 1 1 0 0 0 1 1 1 1 0 0 0 1 2 1 1 3 2 5 1 2 3 5


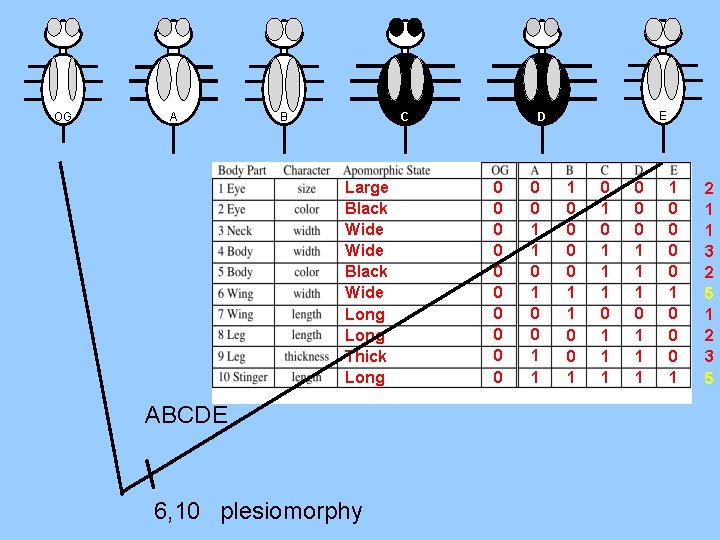
OG A C B Large Black Wide Long Thick Long ABCDE 6, 10 plesiomorphy E D 0 0 0 1 1 0 0 1 1 0 0 1 0 1 1 1 0 0 0 1 1 1 1 0 0 0 1 2 1 1 3 2 5 1 2 3 5

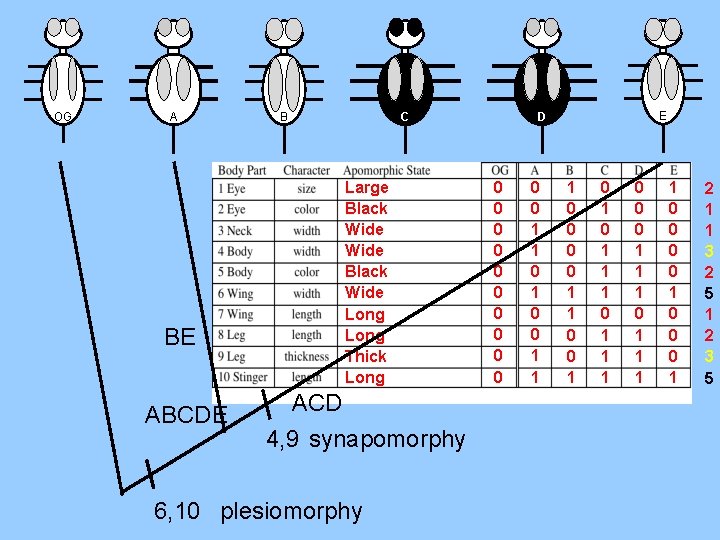
OG A BE ABCDE C B Large Black Wide Long Thick Long ACD 4, 9 synapomorphy 6, 10 plesiomorphy E D 0 0 0 1 1 0 0 1 1 0 0 1 0 1 1 1 0 0 0 1 1 1 1 0 0 0 1 2 1 1 3 2 5 1 2 3 5

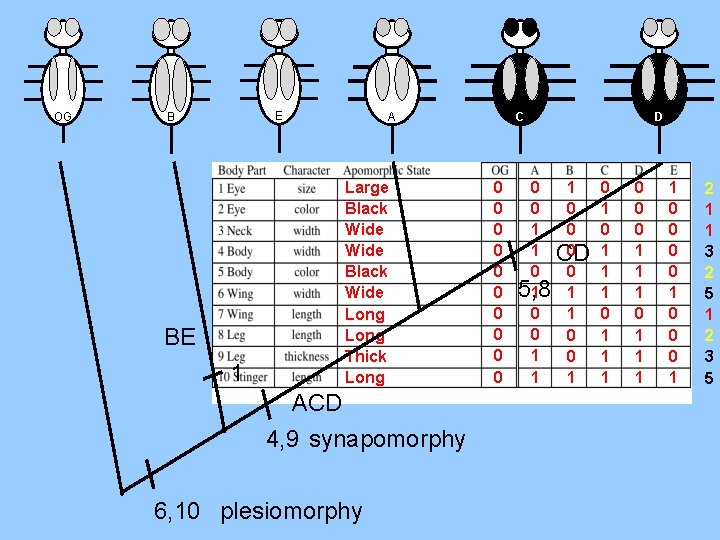
OG E B BE 1 C A Large Black Wide Long Thick Long ACD 4, 9 synapomorphy 6, 10 plesiomorphy 0 0 0 1 1 0 5, 8 1 0 0 1 1 D 1 0 0 0 CD 0 1 1 0 0 1 0 1 1 1 0 0 0 1 1 1 1 0 0 0 1 2 1 1 3 2 5 1 2 3 5

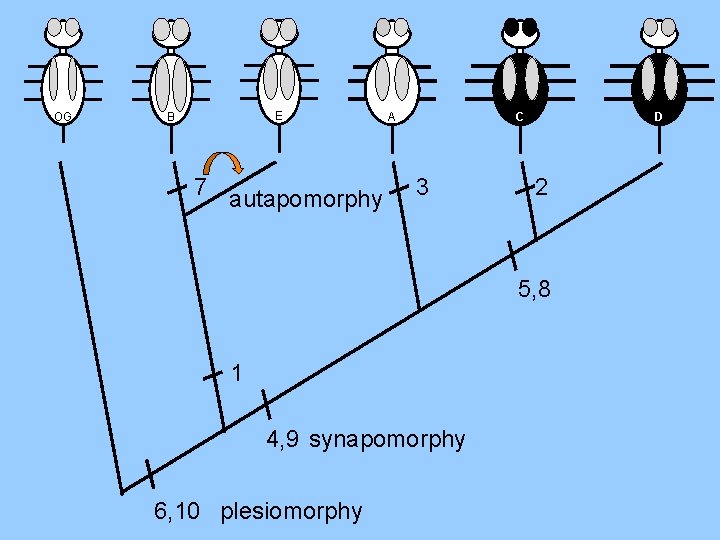
OG E B Large 7 autapomorphy Black BE 1 C A 3 Wide Black Wide Long Thick Long 4, 9 synapomorphy 6, 10 plesiomorphy 0 0 02 0 1 1 0 5, 8 1 0 0 1 1 D 1 0 0 0 CD 0 1 1 0 0 1 0 1 1 1 0 0 0 1 1 1 1 0 0 0 1 2 1 1 3 2 5 1 2 3 5

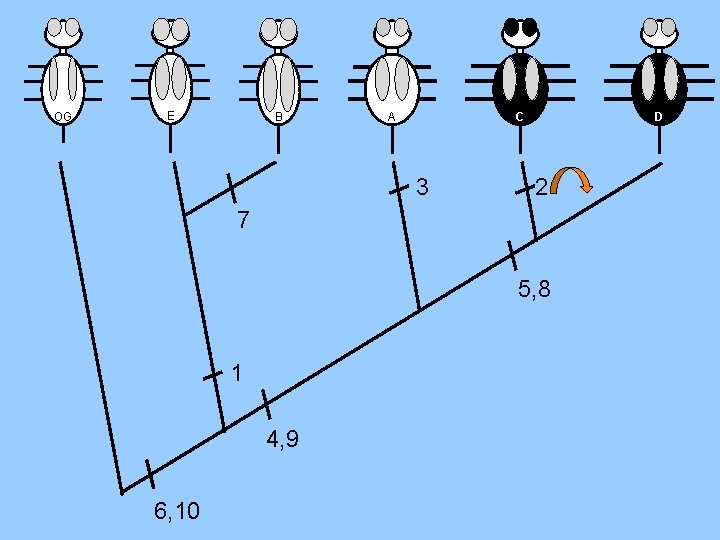
OG E B 7 autapomorphy C A 3 D 2 5, 8 1 4, 9 synapomorphy 6, 10 plesiomorphy

OG E B C A 3 D 2 7 5, 8 1 4, 9 6, 10

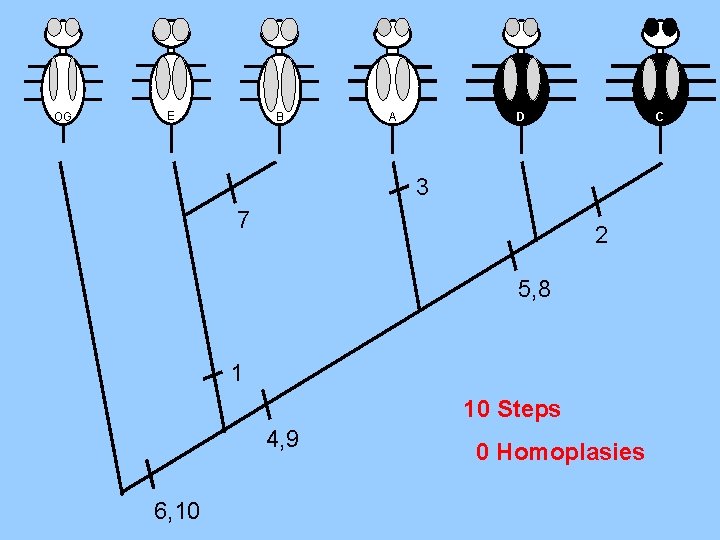
OG E B A C D 3 7 2 5, 8 1 10 Steps 4, 9 6, 10 0 Homoplasies

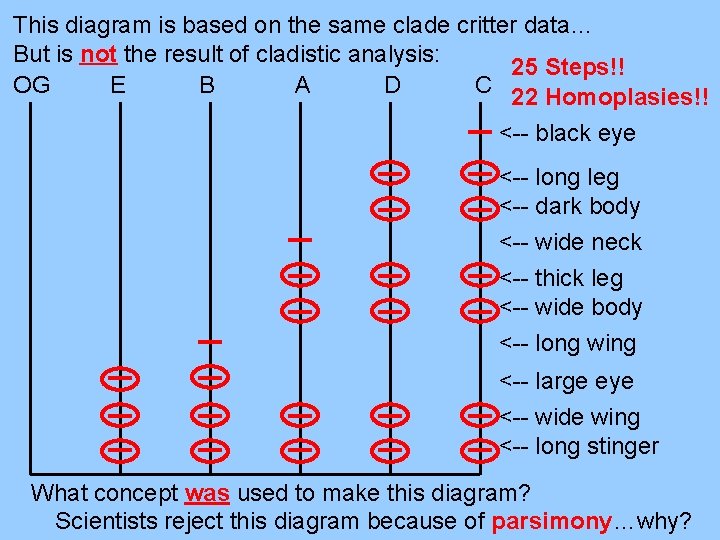
This diagram is based on the same clade critter data… But is not the result of cladistic analysis: 25 Steps!! OG E B A D C 22 Homoplasies!! <-- black eye <-- long leg <-- dark body <-- wide neck <-- thick leg <-- wide body <-- long wing <-- large eye <-- wide wing <-- long stinger What concept was used to make this diagram? Scientists reject this diagram because of parsimony…why?