DOMAINS OF LIFE INTRO 1 EUKARYA EUKARYOTES TRUE

![3 EUKARYOTES ARCHEOBACTERIA EUBACTERIA COMMON ANCESTOR [PROKARYOTIC] 3 EUKARYOTES ARCHEOBACTERIA EUBACTERIA COMMON ANCESTOR [PROKARYOTIC]](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-3.jpg)


![GENUS & SPECIES Staphylococcus aureus YELLOW COLONIES, PATHOGEN [BOILS, PIMPLES, BLOOD INFECTIONS, PNEUMONIA] Staphylococcus GENUS & SPECIES Staphylococcus aureus YELLOW COLONIES, PATHOGEN [BOILS, PIMPLES, BLOOD INFECTIONS, PNEUMONIA] Staphylococcus](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-8.jpg)

![10 MEDIA CAN BE LIQUID OR SOLID [SEMI-SOLID] LIQUID SOLUTIONS SEMI-SOLID - ADD A 10 MEDIA CAN BE LIQUID OR SOLID [SEMI-SOLID] LIQUID SOLUTIONS SEMI-SOLID - ADD A](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-10.jpg)
![STAINS SIMPLE FIX CELLS ON SLIDE ADD DYE [CRYSTAL VIOLET] ALL CELLS TAKE UP STAINS SIMPLE FIX CELLS ON SLIDE ADD DYE [CRYSTAL VIOLET] ALL CELLS TAKE UP](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-13.jpg)

- Slides: 14

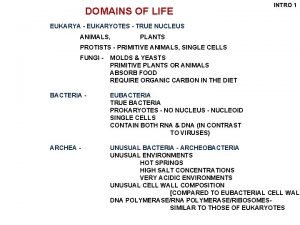
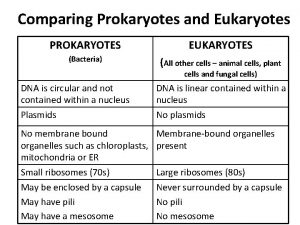
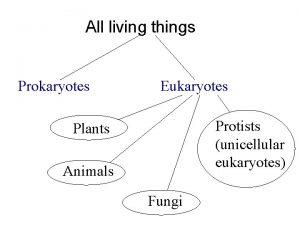
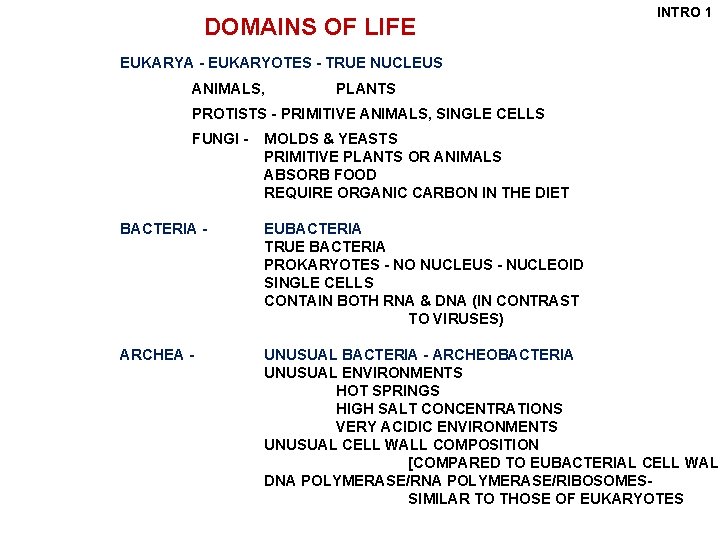
DOMAINS OF LIFE INTRO 1 EUKARYA - EUKARYOTES - TRUE NUCLEUS ANIMALS, PLANTS PROTISTS - PRIMITIVE ANIMALS, SINGLE CELLS FUNGI - MOLDS & YEASTS PRIMITIVE PLANTS OR ANIMALS ABSORB FOOD REQUIRE ORGANIC CARBON IN THE DIET BACTERIA - EUBACTERIA TRUE BACTERIA PROKARYOTES - NO NUCLEUS - NUCLEOID SINGLE CELLS CONTAIN BOTH RNA & DNA (IN CONTRAST TO VIRUSES) ARCHEA - UNUSUAL BACTERIA - ARCHEOBACTERIA UNUSUAL ENVIRONMENTS HOT SPRINGS HIGH SALT CONCENTRATIONS VERY ACIDIC ENVIRONMENTS UNUSUAL CELL WALL COMPOSITION [COMPARED TO EUBACTERIAL CELL WALL DNA POLYMERASE/RIBOSOMESSIMILAR TO THOSE OF EUKARYOTES

2 PHYLOGENY – BASED ON DEGREE OF DNA NUCLEOTIDE SEQUENCE SIMILARITY – ALSO CALLED HOMOLOGY DURING EVOLUTION – MUTATIONS OCCUR, THE NUMBER OF MUTATIONS WITHIN A CHROMOSOME (OR GENE) INCREASES WITH TIME. THEREFORE, TWO ORGANISMS WHICH SEPARATED FROM A COMMON ANCESTOR: LONG AGO WILL HAVE HAD MANY DIFFERENT MUTATIONS AND THE SEQUENCES OF THE GENES WILL HAVE A LOW DEGREE OF SIMILARITY (HOMOLOGY). THOSE ORGANISMS ARE NOT CLOSELY RELATED. SHORT TIME AGO: WILL HAVE HAD RELATIVELY FEW DIFFERENT MUTATIONS AND THE SEQUENCES OF THE GENES WILL HAVE A HIGH DEGREE OF SIMILARITY - THOSE ORGANISMS ARE CLOSELY RELATED
![3 EUKARYOTES ARCHEOBACTERIA EUBACTERIA COMMON ANCESTOR PROKARYOTIC 3 EUKARYOTES ARCHEOBACTERIA EUBACTERIA COMMON ANCESTOR [PROKARYOTIC]](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-3.jpg)
3 EUKARYOTES ARCHEOBACTERIA EUBACTERIA COMMON ANCESTOR [PROKARYOTIC]

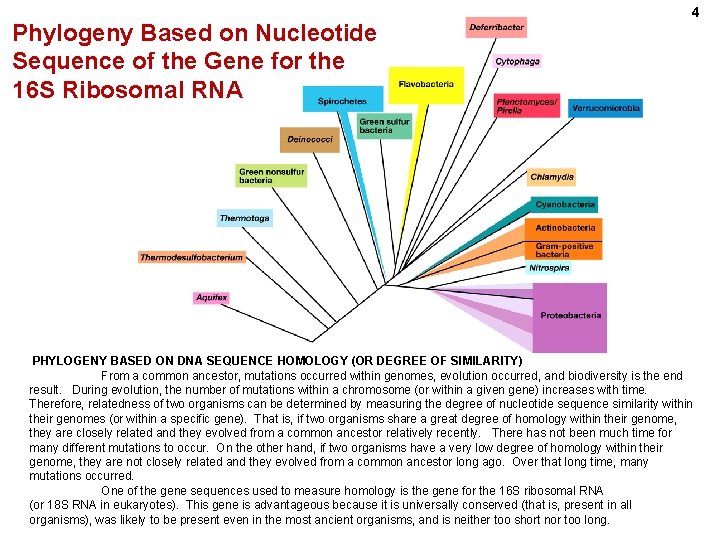
Phylogeny Based on Nucleotide Sequence of the Gene for the 16 S Ribosomal RNA 4 PHYLOGENY BASED ON DNA SEQUENCE HOMOLOGY (OR DEGREE OF SIMILARITY) From a common ancestor, mutations occurred within genomes, evolution occurred, and biodiversity is the end result. During evolution, the number of mutations within a chromosome (or within a given gene) increases with time. Therefore, relatedness of two organisms can be determined by measuring the degree of nucleotide sequence similarity within their genomes (or within a specific gene). That is, if two organisms share a great degree of homology within their genome, they are closely related and they evolved from a common ancestor relatively recently. There has not been much time for many different mutations to occur. On the other hand, if two organisms have a very low degree of homology within their genome, they are not closely related and they evolved from a common ancestor long ago. Over that long time, many mutations occurred. One of the gene sequences used to measure homology is the gene for the 16 S ribosomal RNA (or 18 S RNA in eukaryotes). This gene is advantageous because it is universally conserved (that is, present in all organisms), was likely to be present even in the most ancient organisms, and is neither too short nor too long.

Aquifex: Most ancient Hyperthermophile Chemolithotrophic Spirochetes: Flexible coils Treponema pallidum Borrelia burgdorferi Green Sulfur: Anoxygenic photosynthesis H 2 S S 0 SO 4 Chlamydia: Obligate intracellular bacteria Chlamydia trachomatis Cyanobacteria: Blue green bacteria Oxygenic photosynthesis; First O 2 - evolving creatures on earth Atmosphere converted from anoxic to oxic (Synechococcus) Gram positive: Staphylococcus Bacillus Proteobacteria: Gram negative rods and cocci Most common in medicine, agriculture, industry Phenotypes diverse e. g. Enteric Pseudomonas, N 2 -fixing Azotobacter 5

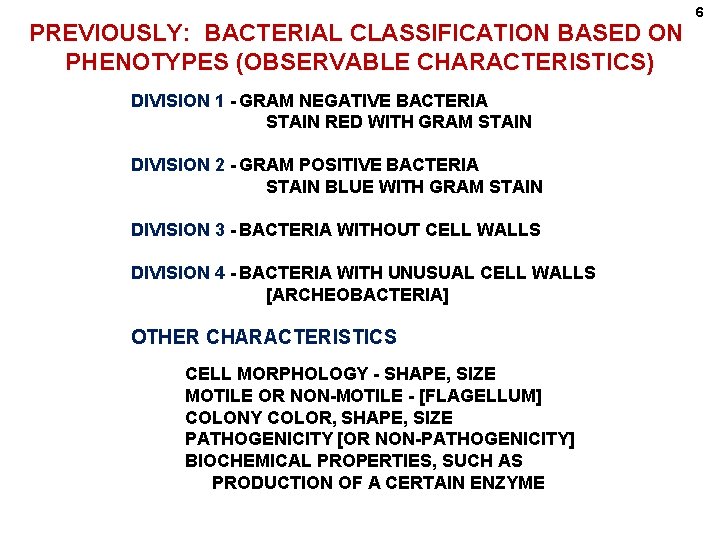
PREVIOUSLY: BACTERIAL CLASSIFICATION BASED ON PHENOTYPES (OBSERVABLE CHARACTERISTICS) DIVISION 1 - GRAM NEGATIVE BACTERIA STAIN RED WITH GRAM STAIN DIVISION 2 - GRAM POSITIVE BACTERIA STAIN BLUE WITH GRAM STAIN DIVISION 3 - BACTERIA WITHOUT CELL WALLS DIVISION 4 - BACTERIA WITH UNUSUAL CELL WALLS [ARCHEOBACTERIA] OTHER CHARACTERISTICS CELL MORPHOLOGY - SHAPE, SIZE MOTILE OR NON-MOTILE - [FLAGELLUM] COLONY COLOR, SHAPE, SIZE PATHOGENICITY [OR NON-PATHOGENICITY] BIOCHEMICAL PROPERTIES, SUCH AS PRODUCTION OF A CERTAIN ENZYME 6

EXAMPLES DIVISION 1 - GRAM NEGATIVE BACTERIA SECTION 1 - SPIROCHETES - SPIRAL SHAPE, FLEXIBLE Treponema pallidum SYPHILIS DIVISION 2 - GRAM POSITIVE BACTERIA SECTION 16 - MYCOBACTERIA - RODS, THICK WAXY CELL WALLS, RESISTANT TO DRYING Mycobacterium tuberculosis TUBERCULOSIS SECTION 12 - SPHERES [COCCI, COCCUS] FAMILY 1 MICROCOCCACEAE GRAM POSITIVE SPHERES, GROW IN: IRREGULAR CLUMPS, or PACKETS OF FOUR, or PACKETS OF EIGHT GENUS - Staphylococcus - GRAM POSITIVE SPHERES, IRREGULAR CLUMPS 7
![GENUS SPECIES Staphylococcus aureus YELLOW COLONIES PATHOGEN BOILS PIMPLES BLOOD INFECTIONS PNEUMONIA Staphylococcus GENUS & SPECIES Staphylococcus aureus YELLOW COLONIES, PATHOGEN [BOILS, PIMPLES, BLOOD INFECTIONS, PNEUMONIA] Staphylococcus](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-8.jpg)
GENUS & SPECIES Staphylococcus aureus YELLOW COLONIES, PATHOGEN [BOILS, PIMPLES, BLOOD INFECTIONS, PNEUMONIA] Staphylococcus epidermidis WHITE COLONIES, GENERALLY NON-PATHOGENIC Streptococcus pyogenes IMPORTANT PATHOGEN, GRAM POSITIVE, SPHERES, GROW IN CHAINS “PYOGENES” - PRODUCING PUS STREP THROAT - RHEUMATIC FEVER, HEART VALVE DAMAGE TOXIC SHOCK SYNDROME 8

9 COMPOSITION OF MEDIA ENRICHED / COMPLEX MEDIA EXTRACT OF BEEF MUSCLE, HEART, BRAIN EXTRACT OF YEAST CELLS [EXTRACT = EVERYTHING DISSOLVING IN WATER] MILK PROTEIN HYDROLYSATE [HYDROLYSATE = MACROMOLECULES BROKEN DOWN INTO LOW MOLECULAR WEIGHT, SOLUBLE COMPONENTS, SUCH AS AMINO ACIDS] DEFINED - EXACTLY KNOWN COMPOSITION GLUCOSE - CARBON / ENERGY SOURCE NITROGEN - NH 4 Cl SULFUR - Mg. SO 4 PHOSPHOROUS - Na 2 HPO 4 COMMON MINERALS Mg++ K+ Na+ Cl. TRACE MINERALS Fe+++ Ca++ WATER
![10 MEDIA CAN BE LIQUID OR SOLID SEMISOLID LIQUID SOLUTIONS SEMISOLID ADD A 10 MEDIA CAN BE LIQUID OR SOLID [SEMI-SOLID] LIQUID SOLUTIONS SEMI-SOLID - ADD A](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-10.jpg)
10 MEDIA CAN BE LIQUID OR SOLID [SEMI-SOLID] LIQUID SOLUTIONS SEMI-SOLID - ADD A GELLING AGENT - AGAR: MELTS AT 100ºC [WHEN THE MEDIUM IS HEATED FOR STERILIZATION] SOLIDIFIES AGAIN AT 41ºC [WHEN THE MEDIUM COOLS] PETRI PLATE

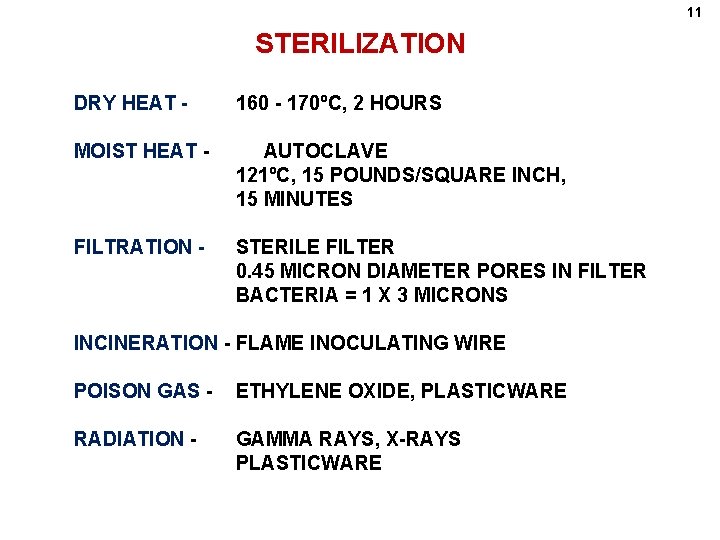
11 STERILIZATION DRY HEAT - 160 - 170ºC, 2 HOURS MOIST HEAT - AUTOCLAVE 121ºC, 15 POUNDS/SQUARE INCH, 15 MINUTES FILTRATION - STERILE FILTER 0. 45 MICRON DIAMETER PORES IN FILTER BACTERIA = 1 X 3 MICRONS INCINERATION - FLAME INOCULATING WIRE POISON GAS - ETHYLENE OXIDE, PLASTICWARE RADIATION - GAMMA RAYS, X-RAYS PLASTICWARE

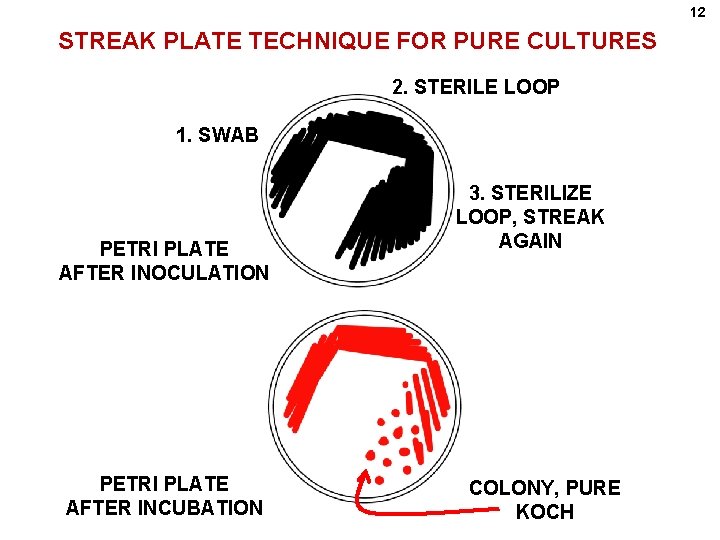
12 STREAK PLATE TECHNIQUE FOR PURE CULTURES 2. STERILE LOOP 1. SWAB PETRI PLATE AFTER INOCULATION PETRI PLATE AFTER INCUBATION 3. STERILIZE LOOP, STREAK AGAIN COLONY, PURE KOCH
![STAINS SIMPLE FIX CELLS ON SLIDE ADD DYE CRYSTAL VIOLET ALL CELLS TAKE UP STAINS SIMPLE FIX CELLS ON SLIDE ADD DYE [CRYSTAL VIOLET] ALL CELLS TAKE UP](https://slidetodoc.com/presentation_image/2a2006cf6a4d9361bae9b9bf1a54375c/image-13.jpg)
STAINS SIMPLE FIX CELLS ON SLIDE ADD DYE [CRYSTAL VIOLET] ALL CELLS TAKE UP DYE WASH AWAY EXCESS DYE NEGATIVE STAIN COLORS BACKGROUND, CELLS APPEAR WHITE GRAM STAIN FIX CELLS CRYSTAL VIOLET PLUS IODINE [ALL CELLS STAIN VIOLET] WASH WITH ALCOHOL [ALCOHOL REMOVES DYE FROM GRAM NEGATIVE CELLS] ADD SAFRANIN - COUNTER STAIN TO STAIN GRAM NEGATIVE CELLS RED ACID FAST FIX CELLS DYE WITH HEAT DECOLORIZE WITH HEAT AND ALCOHOL MOST CELLS ARE NOW COLORLESS; MYCOBACTERIA RETAIN RED DYE 13

14 FLUORESCENCE MICROSCOPY AFFIX CELLS ONTO SLIDE REACT WITH FLUORESCENT REAGENT WHICH IS SPECIFIC FOR THOSE CELLS OBSERVE WITH FLUORESCENT MICROSCOPE
 True life intro
True life intro Domain of life
Domain of life Organizing life's diversity
Organizing life's diversity Three domains of life
Three domains of life Life domains
Life domains Life domains
Life domains True or false true or false
True or false true or false Chapter 2 jesus christ true god and true man
Chapter 2 jesus christ true god and true man Images of unicellular and multicellular organisms
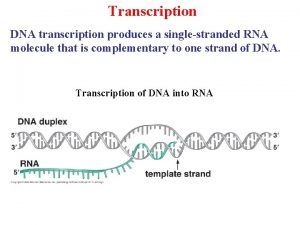
Images of unicellular and multicellular organisms Transcription initiation in eukaryotes
Transcription initiation in eukaryotes Repressible operon
Repressible operon Prokaryotic promoter vs eukaryotic promoter
Prokaryotic promoter vs eukaryotic promoter General characteristics of ascomycota
General characteristics of ascomycota Fine structure analysis of eukaryotes
Fine structure analysis of eukaryotes Translation in bacteria vs eukaryotes
Translation in bacteria vs eukaryotes