DNA Technology Dr Esmaeilzadeh DNA Technology have two



- Slides: 30

DNA Technology Dr. Esmaeilzadeh

DNA Technology have two main parts: ØDNA amplification ØMethods that evaluate DNA

DNA cloning • DNA cloning is the process of making multiple, identical copies of a particular piece of DNA. It can be achieved by two different approuches: • Cell based: Methods that using host cell mechanisms for DNA amplification (In vivo). • Polymerase Chain Reaction=PCR (In vitro).

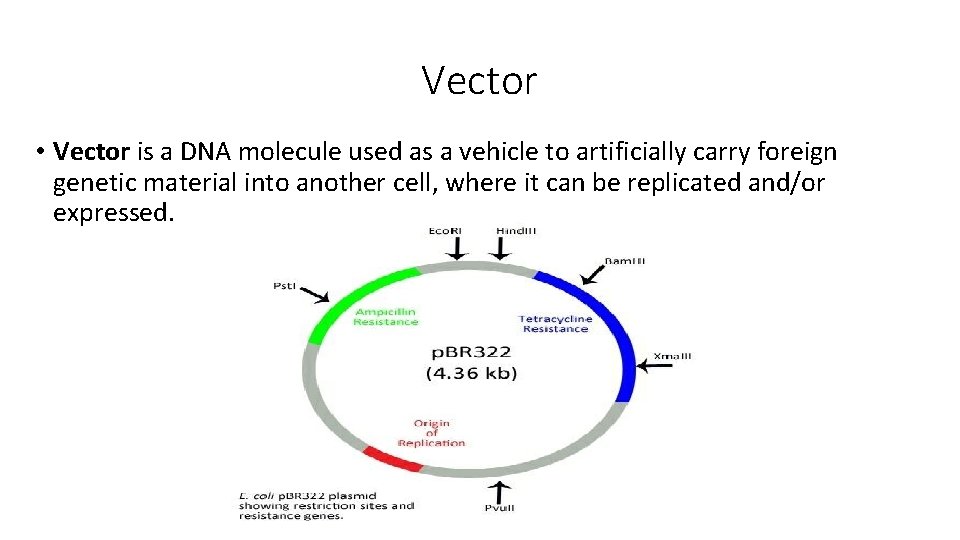
Vector • Vector is a DNA molecule used as a vehicle to artificially carry foreign genetic material into another cell, where it can be replicated and/or expressed.

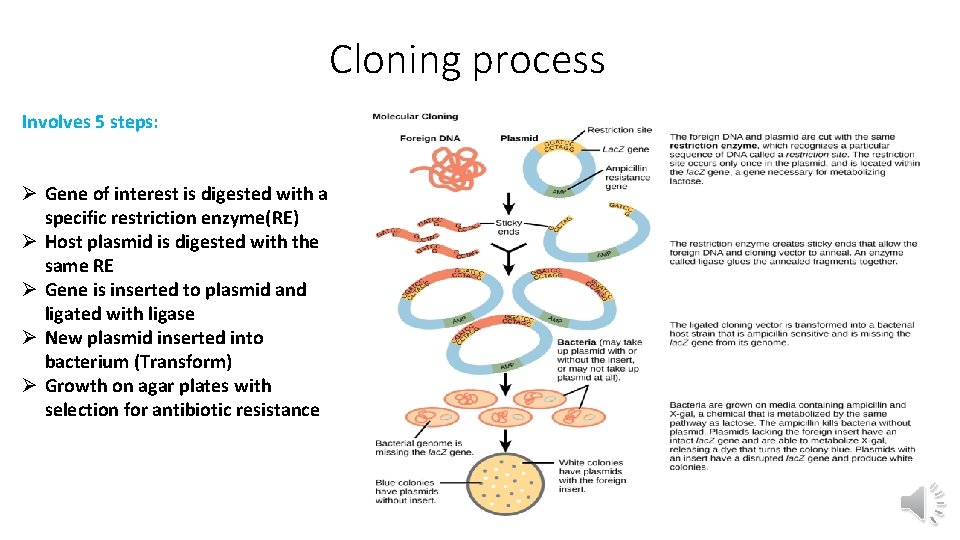
Cloning process Involves 5 steps: Ø Gene of interest is digested with a specific restriction enzyme(RE) Ø Host plasmid is digested with the same RE Ø Gene is inserted to plasmid and ligated with ligase Ø New plasmid inserted into bacterium (Transform) Ø Growth on agar plates with selection for antibiotic resistance

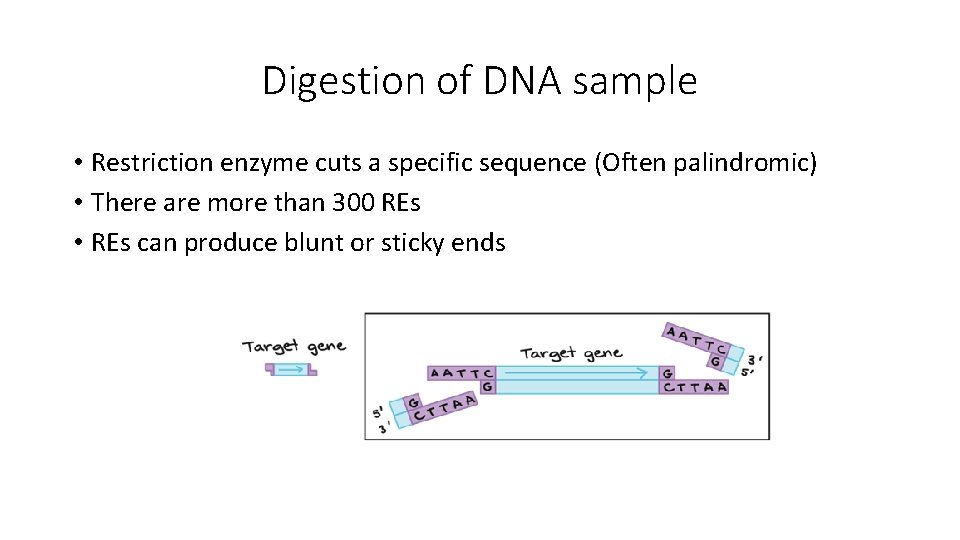
Digestion of DNA sample • Restriction enzyme cuts a specific sequence (Often palindromic) • There are more than 300 REs • REs can produce blunt or sticky ends

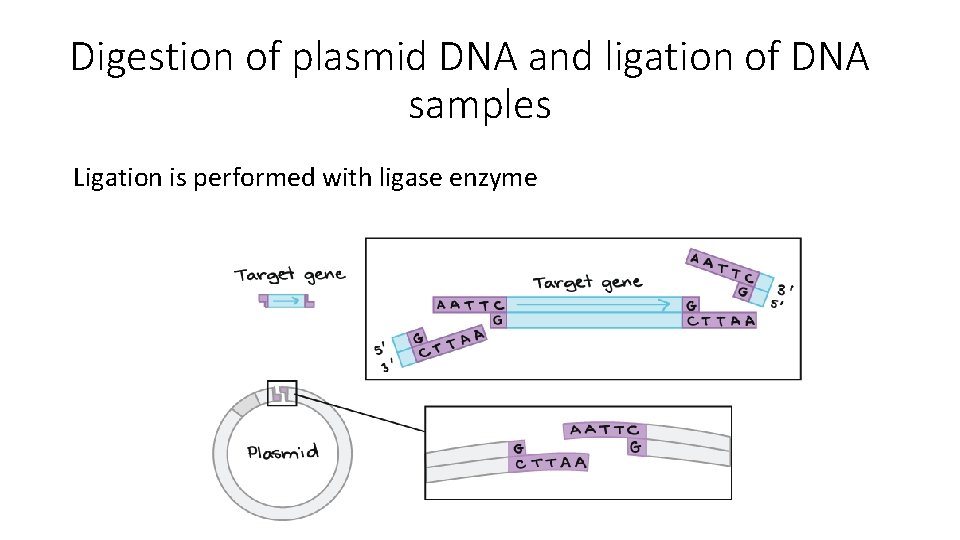
Digestion of plasmid DNA and ligation of DNA samples Ligation is performed with ligase enzyme

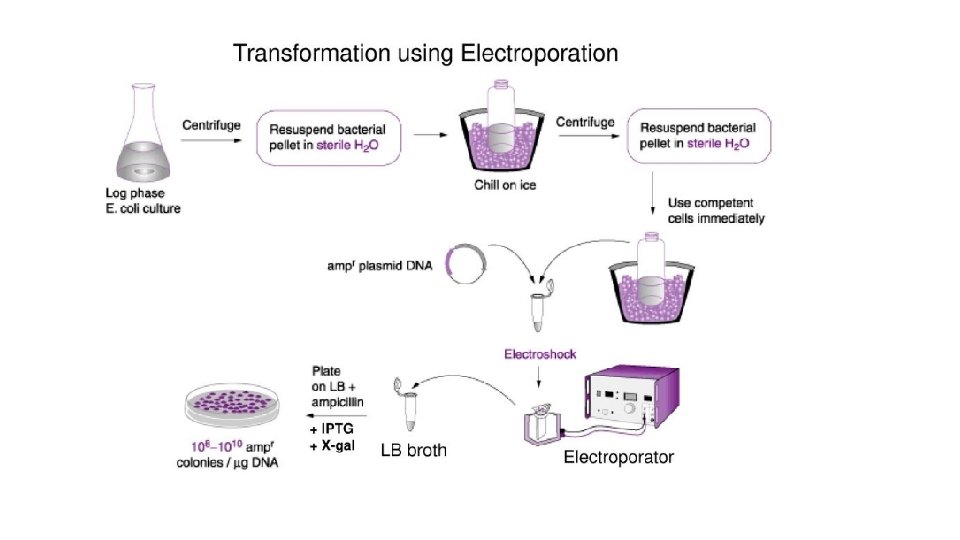
Transformation of ligation products • The process of transferring exogenous DNA into host cells is called transformation. • There are basically two general methods to transforming bacteria: üChemical method using Ca. Cl 2 and heat shock üElectroporation (Based on short pulses of electric charge)

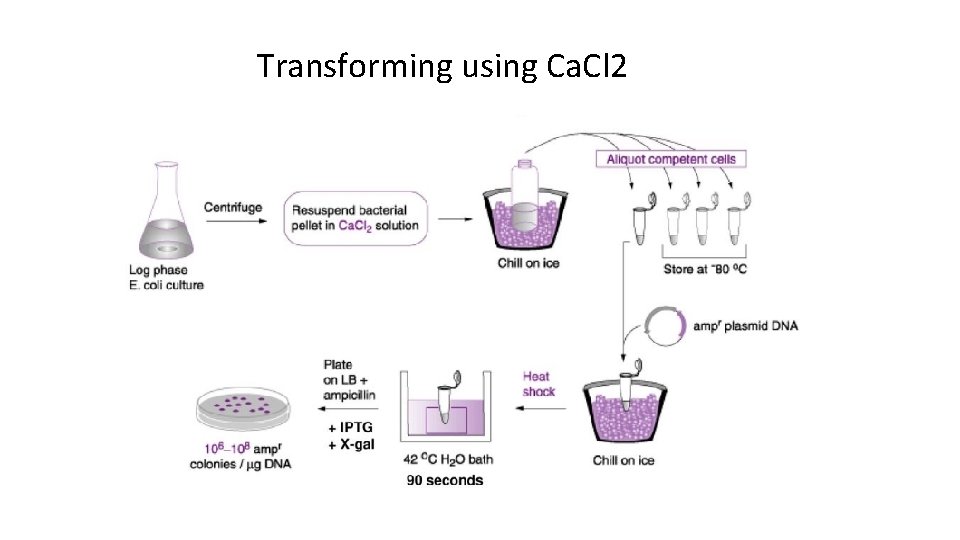
Transforming using Ca. Cl 2


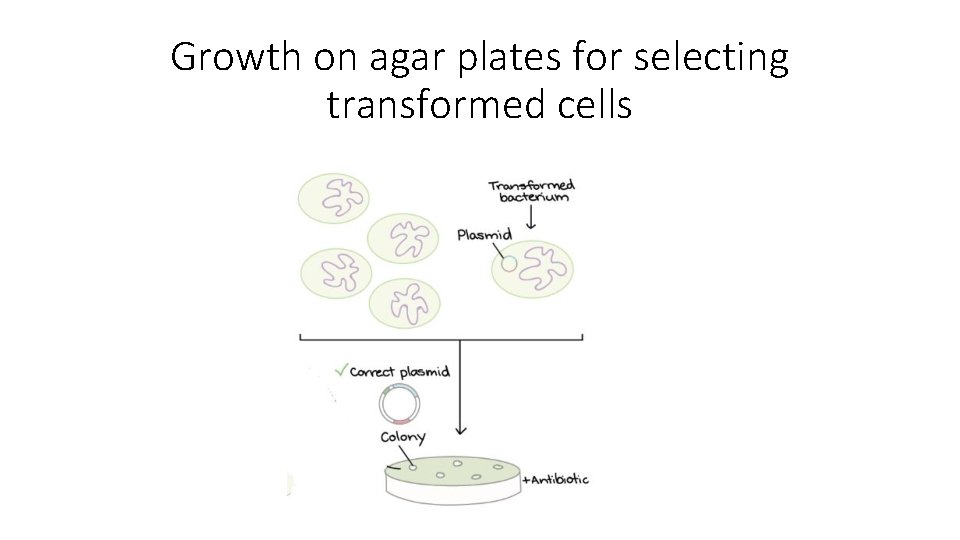
Growth on agar plates for selecting transformed cells

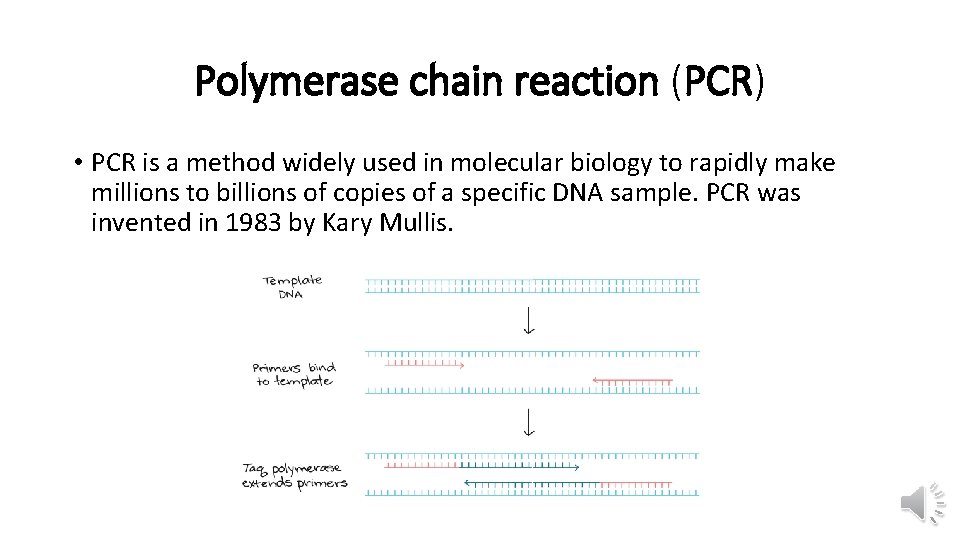
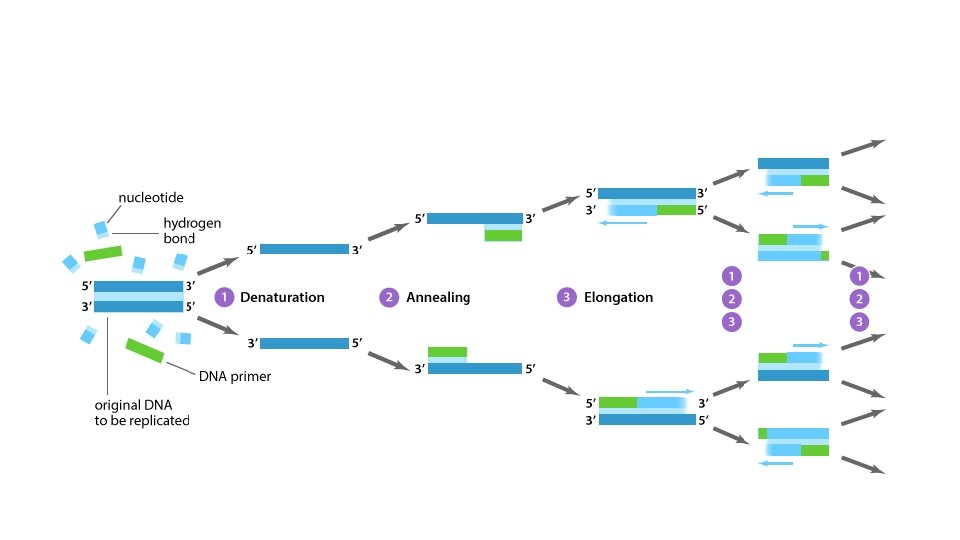
Polymerase chain reaction (PCR) • PCR is a method widely used in molecular biology to rapidly make millions to billions of copies of a specific DNA sample. PCR was invented in 1983 by Kary Mullis.

PCR • There are three main stages: • Denaturing : when the double-stranded template DNA is heated to separate it into two single strands. • Annealing : when the temperature is lowered to enable the DNA primers to attach to the template DNA. • Extending : when the temperature is raised and the new strand of DNA is made by the Taq polymerase enzyme. • These three stages are repeated 20 -40 times, doubling the number of DNA copies each time.


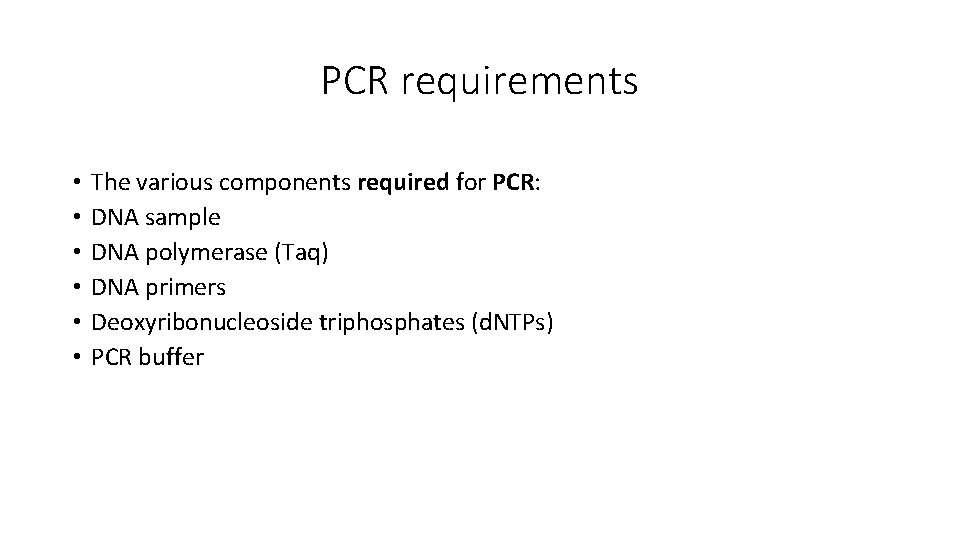
PCR requirements • • • The various components required for PCR: DNA sample DNA polymerase (Taq) DNA primers Deoxyribonucleoside triphosphates (d. NTPs) PCR buffer

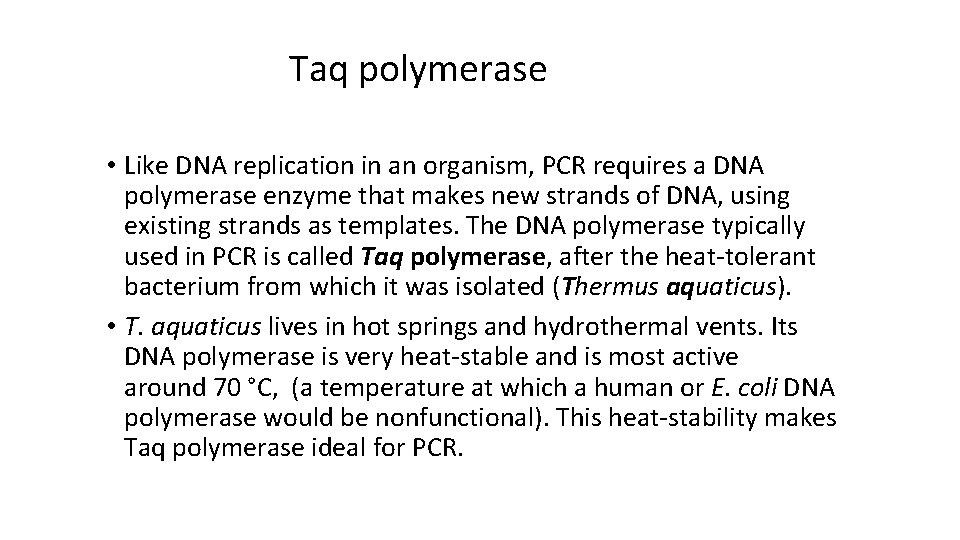
Taq polymerase • Like DNA replication in an organism, PCR requires a DNA polymerase enzyme that makes new strands of DNA, using existing strands as templates. The DNA polymerase typically used in PCR is called Taq polymerase, after the heat-tolerant bacterium from which it was isolated (Thermus aquaticus). • T. aquaticus lives in hot springs and hydrothermal vents. Its DNA polymerase is very heat-stable and is most active around 70 °C, (a temperature at which a human or E. coli DNA polymerase would be nonfunctional). This heat-stability makes Taq polymerase ideal for PCR.

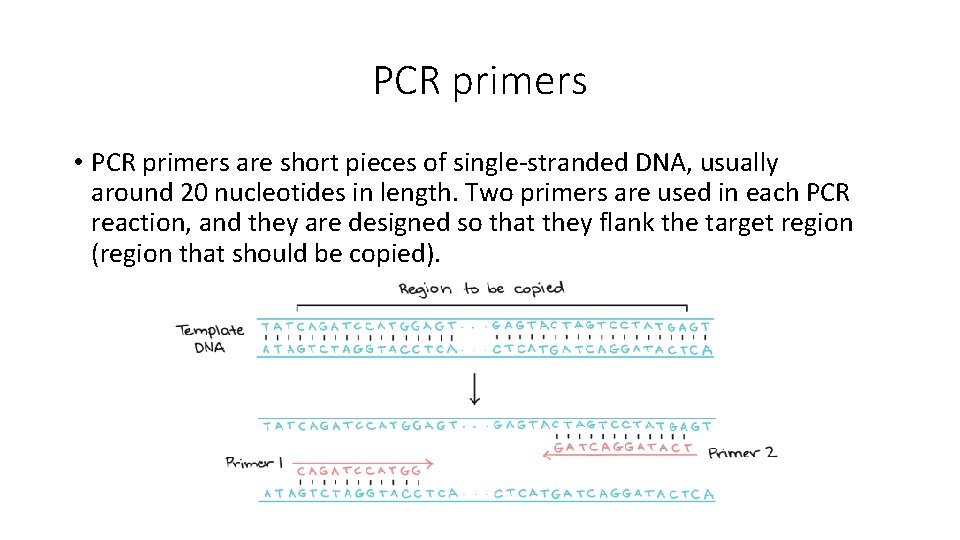
PCR primers • PCR primers are short pieces of single-stranded DNA, usually around 20 nucleotides in length. Two primers are used in each PCR reaction, and they are designed so that they flank the target region (region that should be copied).

Primer designing criteria • Primers should generally have the following properties: • Length of 18 -24 bases • 40 -60% GC content • Start and end with 1 -2 G or C bases • Melting temperature (Tm) of 50 -60°C • Primer pairs should have a Tm within 5°C of each other • Primer pairs should not have complementary regions

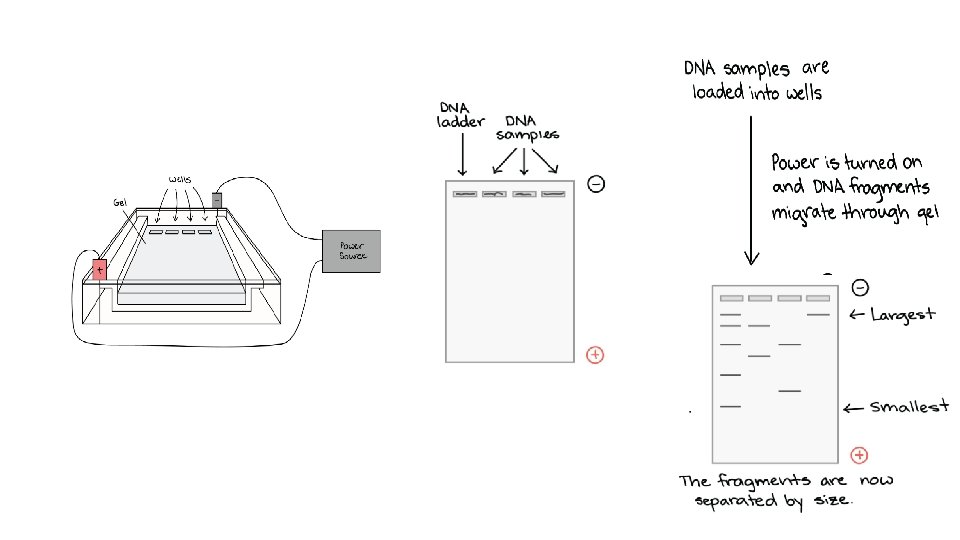
Gel electrophoresis • Gel electrophoresis is a technique used to separate DNA fragments according to their size. üDNA samples are loaded into wells (indentations) at one end of a gel, and an electric current is applied to pull them through the gel. üDNA fragments are negatively charged, so they move towards the positive electrode. Because all DNA fragments have the same amount of charge per mass, small fragments move through the gel faster than large ones. üWhen a gel is stained with a DNA-binding dye, the DNA fragments can be seen as bands, each representing a group of same-sized DNA fragments.


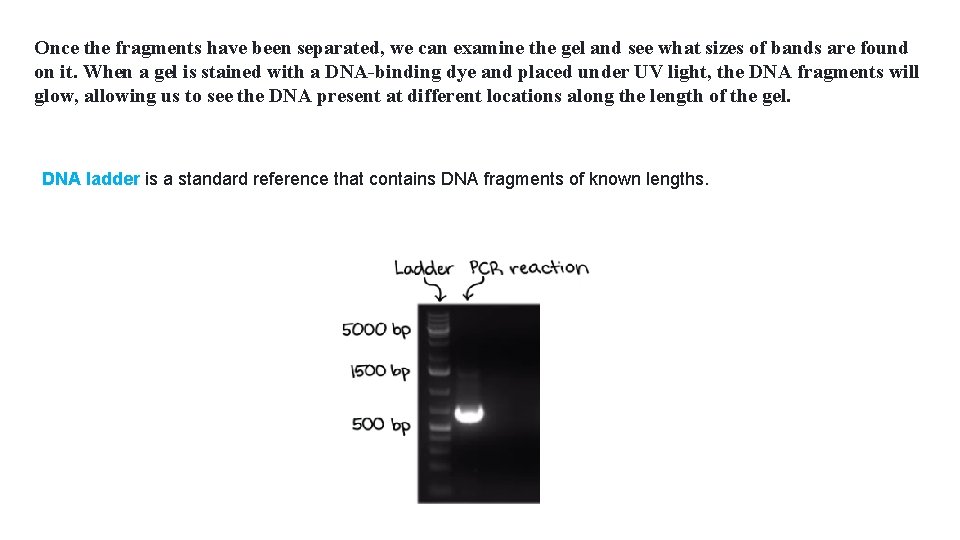
Once the fragments have been separated, we can examine the gel and see what sizes of bands are found on it. When a gel is stained with a DNA-binding dye and placed under UV light, the DNA fragments will glow, allowing us to see the DNA present at different locations along the length of the gel. DNA ladder is a standard reference that contains DNA fragments of known lengths.

Types of polymerase chain reaction-PCR • Several modification of PCR methods have been developed to enhance the utility of this method in diagnostic settings based on their applications. Some of the common types of PCR are: • Real-Time PCR (quantitative PCR or q. PCR) • Multiplex PCR • Nested PCR • ARMS PCR • …

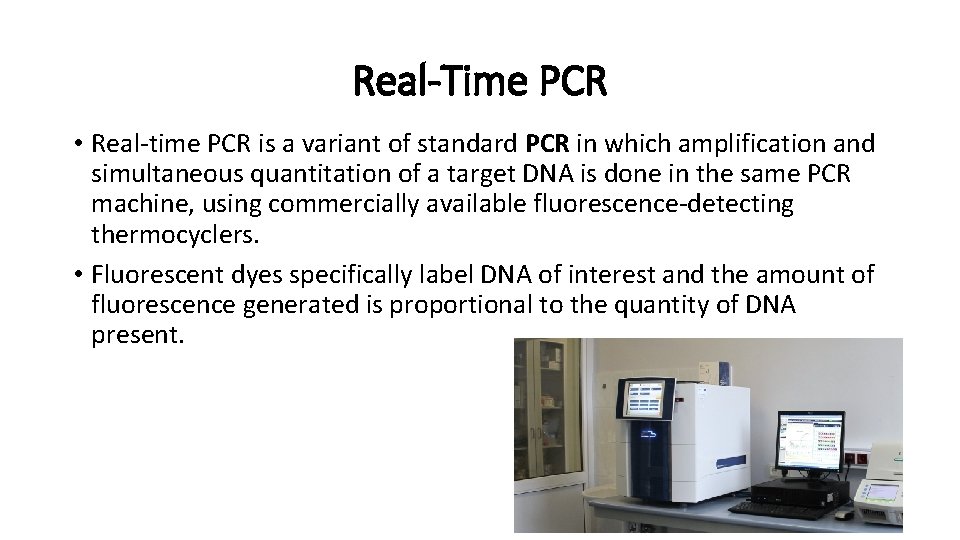
Real-Time PCR • Real-time PCR is a variant of standard PCR in which amplification and simultaneous quantitation of a target DNA is done in the same PCR machine, using commercially available fluorescence-detecting thermocyclers. • Fluorescent dyes specifically label DNA of interest and the amount of fluorescence generated is proportional to the quantity of DNA present.

• Multiplex PCR is a variant of PCR method in which more than one target sequence is amplified using multiple sets of primers within a single PCR mixture. This enables amplification of several gene segments at the same time, instead of specific test runs for each. • Nested PCR is a modification of PCR designed to increase the sensitivity and specificity of the assay reaction. It involves the use of two primer sets directed against the same target and two successive PCR reactions.

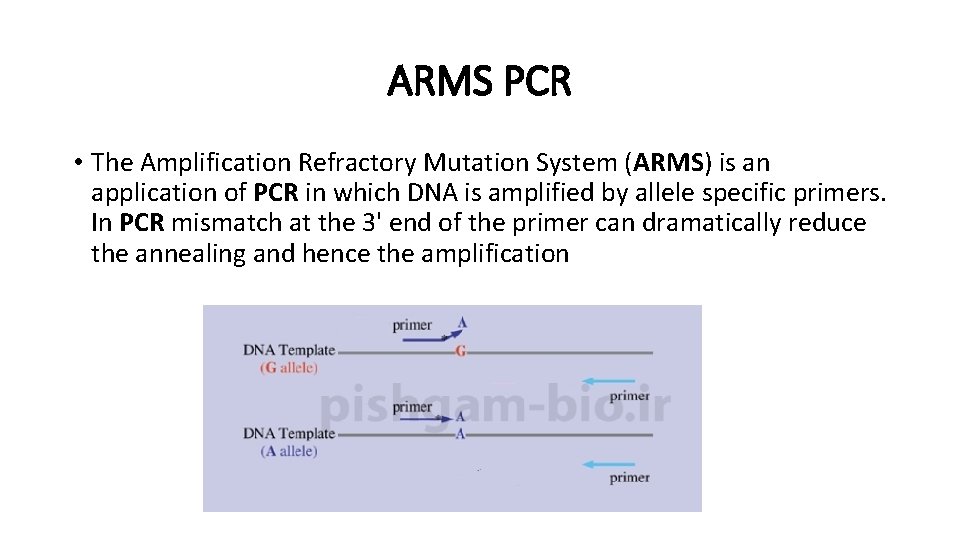
ARMS PCR • The Amplification Refractory Mutation System (ARMS) is an application of PCR in which DNA is amplified by allele specific primers. In PCR mismatch at the 3' end of the primer can dramatically reduce the annealing and hence the amplification

Applications of PCR • Identification and characterization of infectious agents • • Direct detection of microorganisms in patient specimens Identification of microorganisms grown in culture Detection of antimicrobial resistance Investigation of strain relatedness of pathogen of interest • Genetic fingerprinting (forensic application/paternity testing) • Detection of mutation ( investigation of genetic diseases) • Cloning genes • PCR sequencing

DNA Sequencing • DNA sequencing is the process of determining the sequence of nucleotides within DNA. • There are two main types of DNA sequencing: üSanger Sequencing üHigh Throughput Sequencing

Sanger Sequencing • Sanger sequencing is a method of DNA sequencing based on the selective incorporation of chain-terminating dideoxynucleotides by DNA polymerase during in vitro DNA replication. Developed by Frederick Sanger and colleagues in 1977.

High Throughput Sequencing • Next-generation sequencing (NGS), also known as high-throughput sequencing, is the catch-all term used to describe a number of different modern sequencing technologies. These technologies allow for sequencing of DNA and RNA much more quickly and cheaply than the previously used Sanger sequencing, and as such revolutionised the study of genomics and molecular biology.

Good luck!