DNA Structure The Watson Crick structure for DNA











- Slides: 42

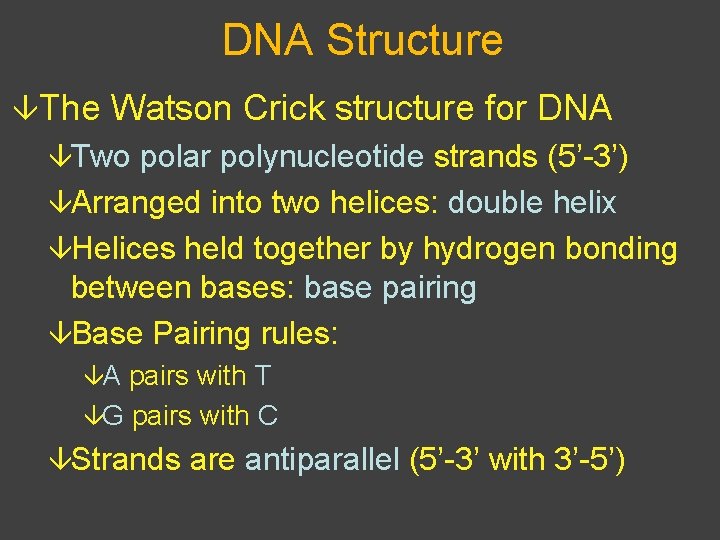
DNA Structure âThe Watson Crick structure for DNA âTwo polar polynucleotide strands (5’-3’) âArranged into two helices: double helix âHelices held together by hydrogen bonding between bases: base pairing âBase Pairing rules: âA pairs with T âG pairs with C âStrands are antiparallel (5’-3’ with 3’-5’)

Imagine a Ladder Then give it a twist

Strands are 5’ end antiparallel H-bonds between O G nitrogenous C bases Base Pairing P O Phosphodiester bond P P O O T A C G T A 3’ end O O O P P P O P 3’ end 5’ end Sugar-phosphate "backbone"

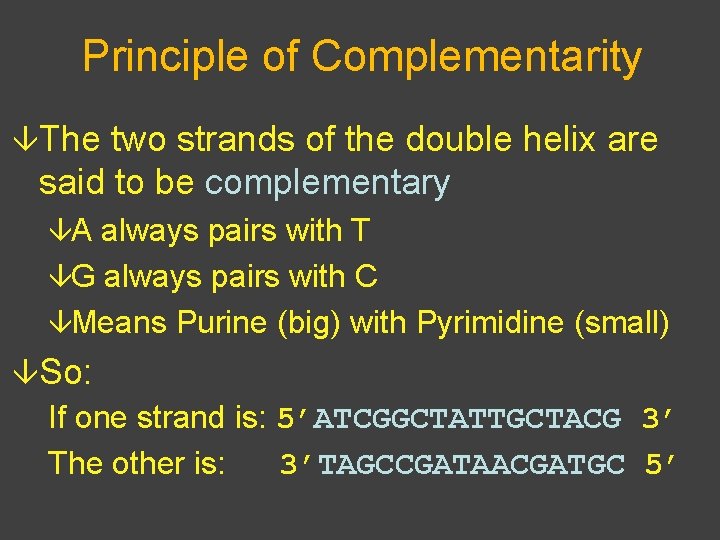
Principle of Complementarity âThe two strands of the double helix are said to be complementary âA always pairs with T âG always pairs with C âMeans Purine (big) with Pyrimidine (small) âSo: If one strand is: 5’ATCGGCTATTGCTACG 3’ The other is: 3’TAGCCGATAACGATGC 5’

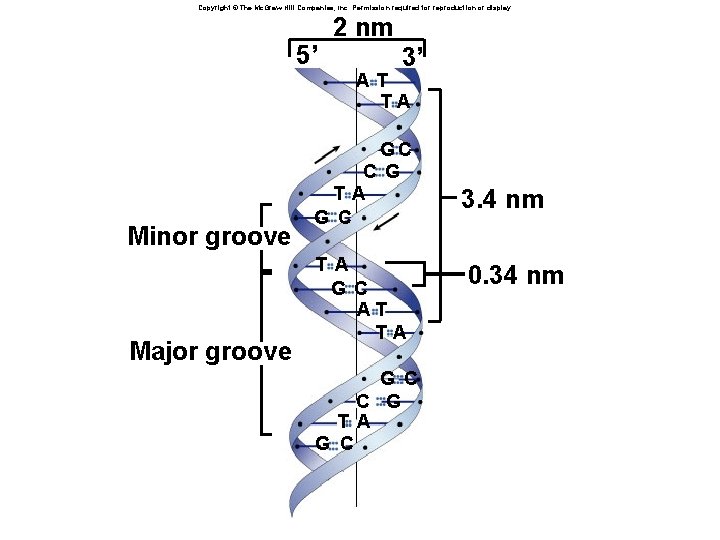
Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. 5’ 2 nm 3’ AT TA Minor groove Major groove GC C G T A G C AT TA G C C G T A G C 3. 4 nm 0. 34 nm

Basic Mechanism of DNA Replication âOpen up helix âCopy each strand âProduces 2 new strands that are each hybrids of new and old strands

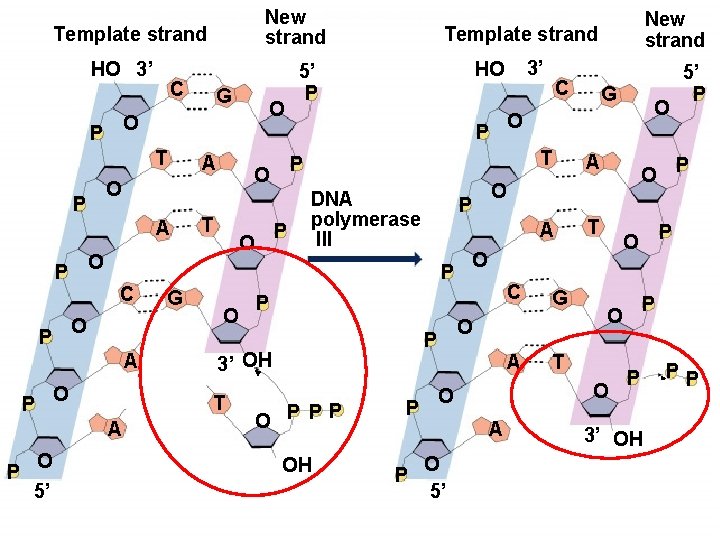
Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. New strand Template strand HO 3’ C A P O O 5’ T P O DNA polymerase III P O O P PP OH P P T A C A A T G A P O T 5’ P P O O 5’ O O O P G O P 3’ OH C O P G 3’ HO 5’ P P O T A P A O C Template strand P T P O O O P P G New strand P 3’ OH PP

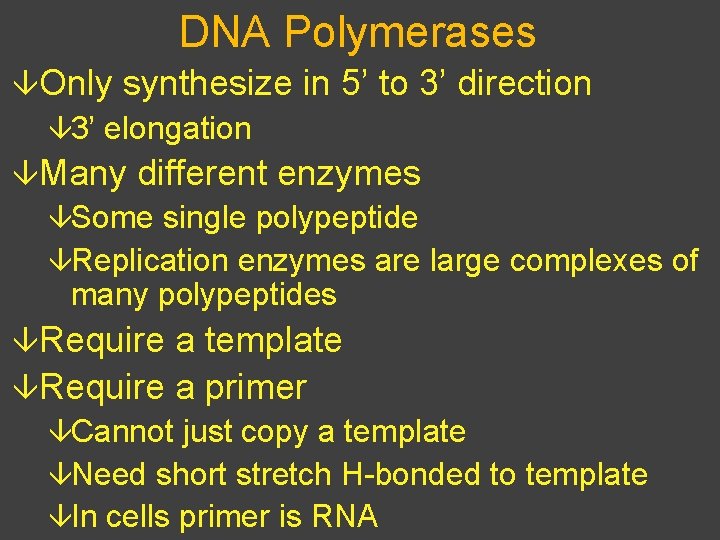
DNA Polymerases âOnly synthesize in 5’ to 3’ direction â 3’ elongation âMany different enzymes âSome single polypeptide âReplication enzymes are large complexes of many polypeptides âRequire a template âRequire a primer âCannot just copy a template âNeed short stretch H-bonded to template âIn cells primer is RNA

Priming of DNA Synthesis

DNA Replication âRequires a number of enzymes that work together âEntire complex is called replisome âEnzyme to open helix âRequires synthesizing RNA primers âPrimers are extended by DNA polymerase âPrimers are then removed âPieces are stitched together âResult is two molecules, each made of template plus newly synthesized strands

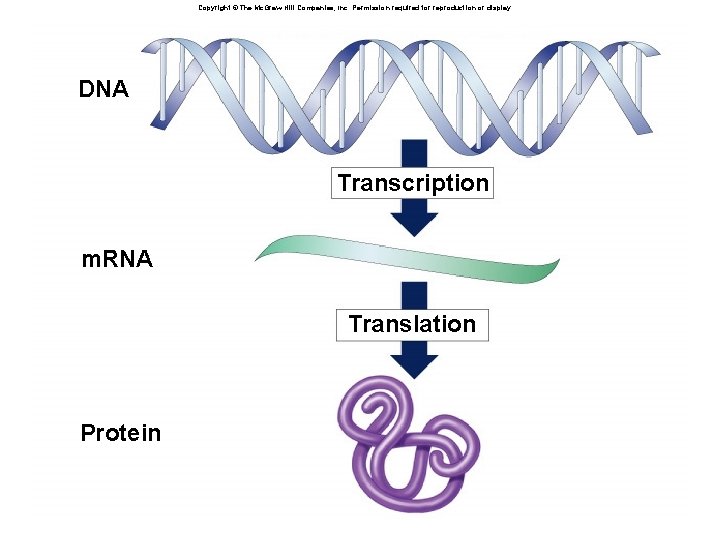
Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. DNA Transcription m. RNA Translation Protein

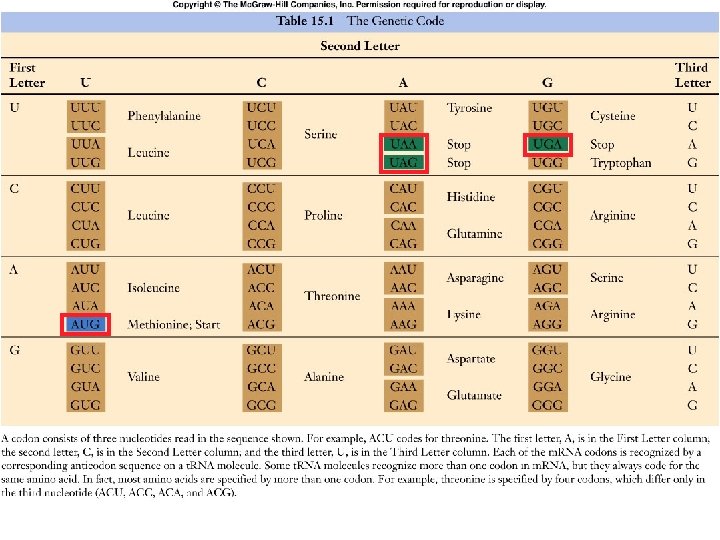
Genetic Code âBefore considering mechanics of gene expression, we need to understand the informatics âRelate information in DNA to information in protein âThe genetic code âCracking the code was a milestone of 20 th century biology âCombination of genetics, biochemistry and organic chemistry

How Many Bases Needed? âDNA “alphabet” to protein“alphabet” âHow many bases needed to specify all amino acids? â 20 amino acids â 4 bases âSo… â 1 base can encode 4 aa’s â 2 bases can encode 16 aa’s â 3 bases can encode 64 aa’s âLater verified experimentally

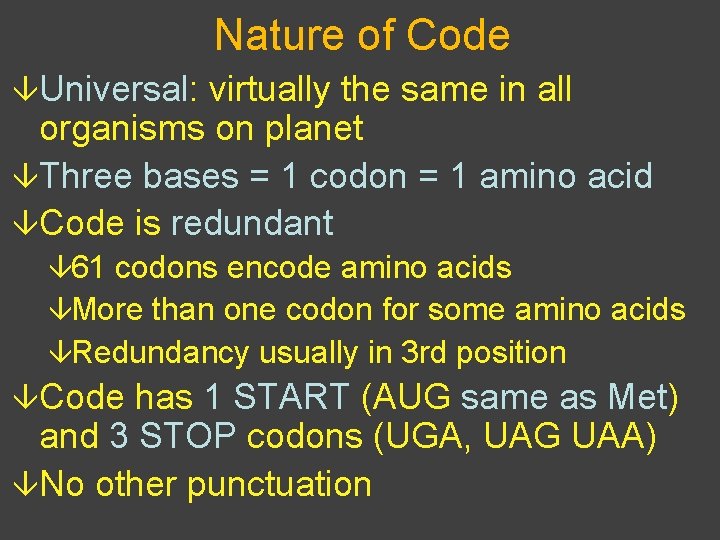
Nature of Code âUniversal: virtually the same in all organisms on planet âThree bases = 1 codon = 1 amino acid âCode is redundant â 61 codons encode amino acids âMore than one codon for some amino acids âRedundancy usually in 3 rd position âCode has 1 START (AUG same as Met) and 3 STOP codons (UGA, UAG UAA) âNo other punctuation

Table 15. 1

Overview of Gene Expression âTranscription: conversion of information in DNA into RNA âRNA: many roles in gene expression as we will see âTemplate âAdapter âRibosome âTranslation or Protein synthesis: information from message —> protein

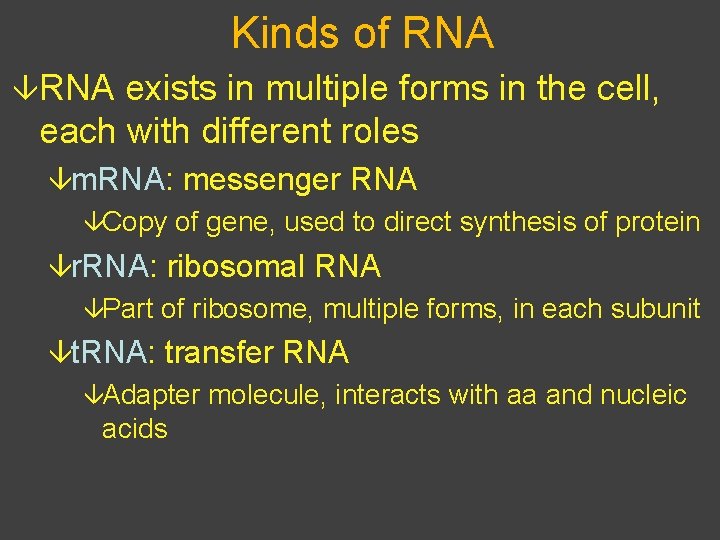
Kinds of RNA âRNA exists in multiple forms in the cell, each with different roles âm. RNA: messenger RNA âCopy of gene, used to direct synthesis of protein âr. RNA: ribosomal RNA âPart of ribosome, multiple forms, in each subunit ât. RNA: transfer RNA âAdapter molecule, interacts with aa and nucleic acids

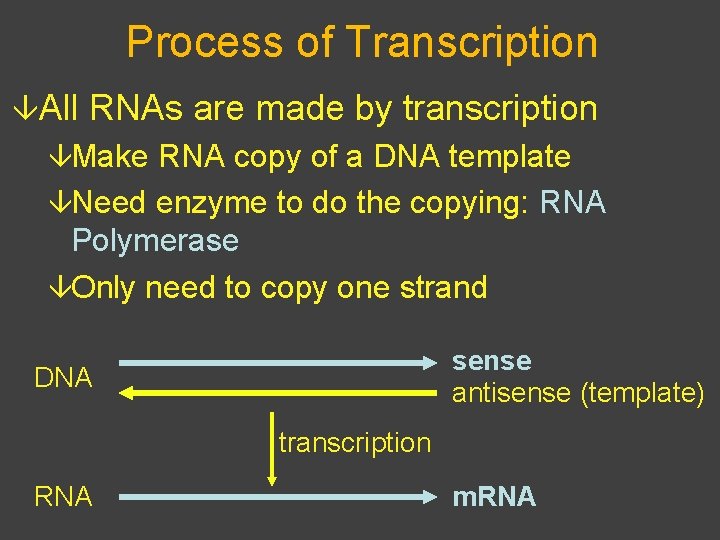
Process of Transcription âAll RNAs are made by transcription âMake RNA copy of a DNA template âNeed enzyme to do the copying: RNA Polymerase âOnly need to copy one strand sense antisense (template) DNA transcription RNA m. RNA

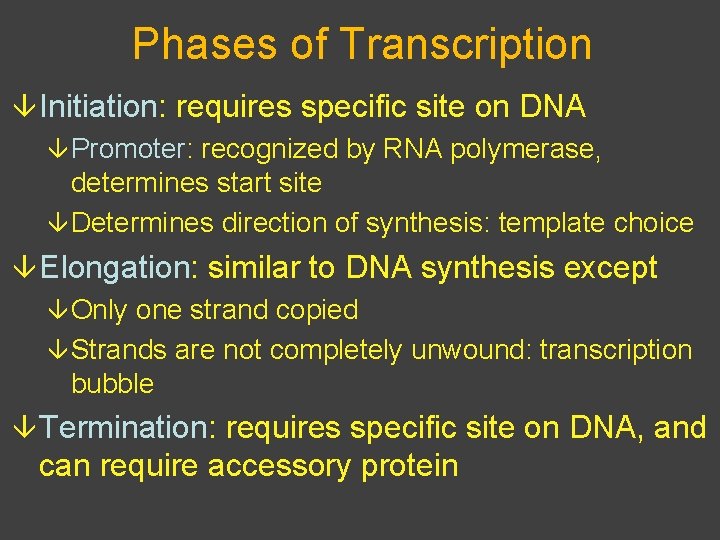
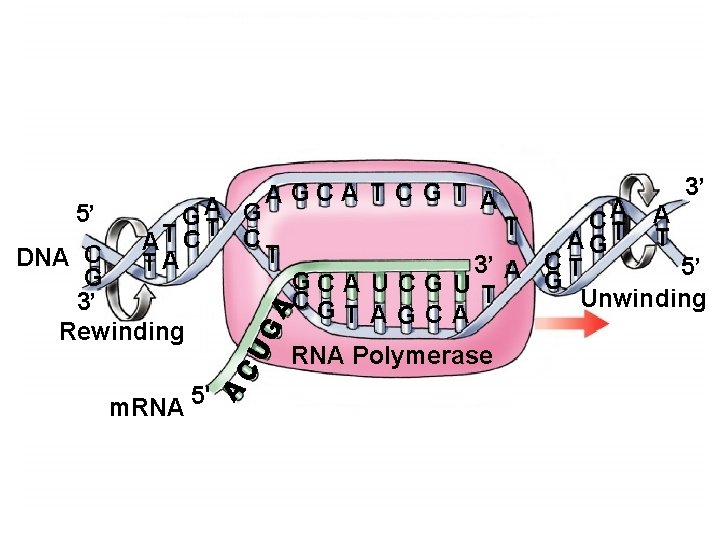
Phases of Transcription â Initiation: requires specific site on DNA â Promoter: recognized by RNA polymerase, determines start site â Determines direction of synthesis: template choice â Elongation: similar to DNA synthesis except â Only one strand copied â Strands are not completely unwound: transcription bubble â Termination: requires specific site on DNA, and can require accessory protein

Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. 5’ A GCA T C G T A G GA T C AT C T TA DNA C G 3’ Rewinding m. RNA A A C T A GT T 3’ A C T 5’ G GC A U C G U T Unwinding CG T AGCA RNA Polymerase 5' 3’

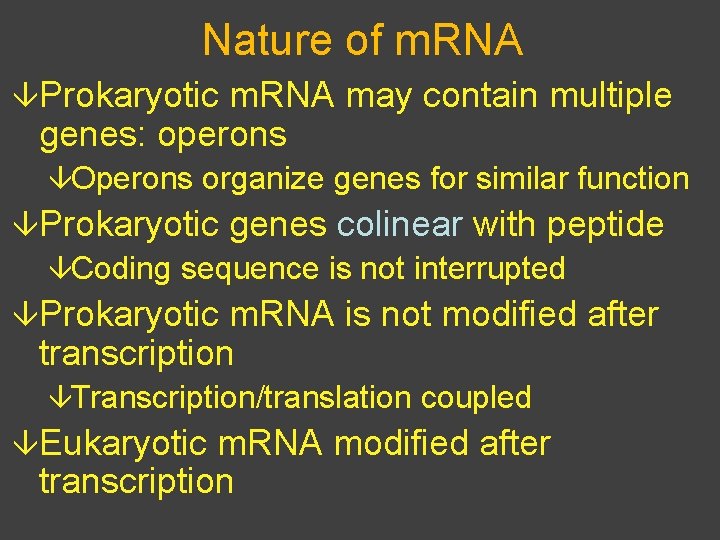
Nature of m. RNA âProkaryotic m. RNA may contain multiple genes: operons âOperons organize genes for similar function âProkaryotic genes colinear with peptide âCoding sequence is not interrupted âProkaryotic m. RNA is not modified after transcription âTranscription/translation coupled âEukaryotic m. RNA modified after transcription

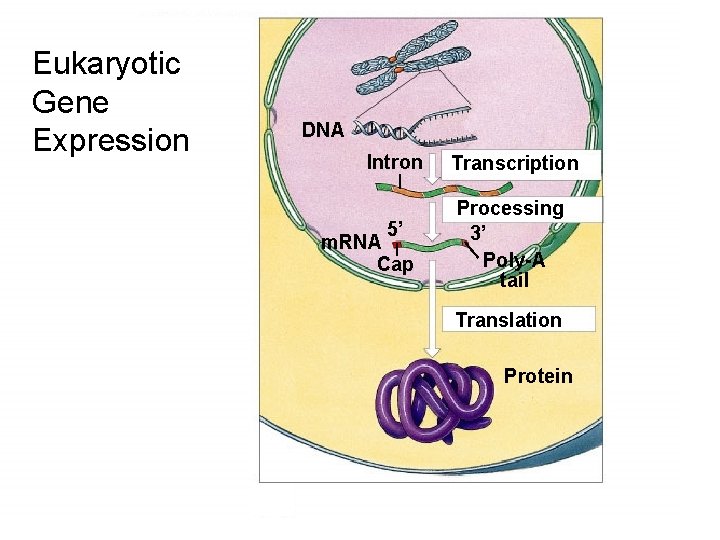
Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. Eukaryotic Gene Expression DNA Intron 5’ m. RNA Cap Transcription Processing 3’ Poly-A tail Translation Protein

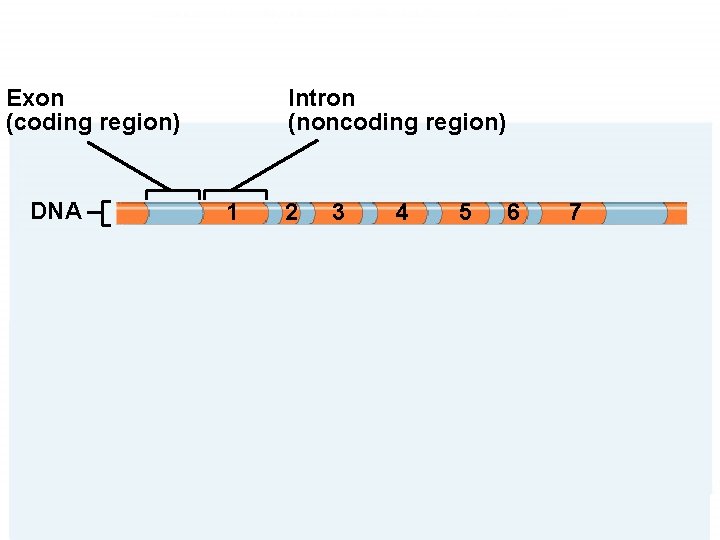
Eukaryotic m. RNA Splicing âEukaryotic genes can be “split” âOrganized into functional (protein coding) and nonfunctional sequences (noncoding âExons: encode protein (expressed) âIntrons: interrupt the exons and are not found in message âSplicing: remove introns connect exons âMust be exact or message will be nonsense

Exon (coding region) DNA Intron (noncoding region) 1 2 3 4 5 6 Transcription Primary RNA transcript Introns are cut out and coding regions are spliced together Mature m. RNA transcript 7

Alternative Splicing âCan splice m. RNA in multiple ways âConfuses nature of the gene: âCan have multiple m. RNA from 1 gene âCan have multiple proteins from 1 gene âCan control type of gene product âCan be tissue specific âCan be sex specific âCan be developmental time specific â 60% of human genes alternatively spliced

Process of Translation âCall this process Protein Synthesis âComponents: âInformation: m. RNA (made from gene) âAdapter molecule/monomer: t. RNA âRibosome: synthetic apparatus, support platform for components âEnergy âTape reading metaphor

Protein Synthesis âBy far the most complex process we will look at in gene expression âAlso the most energy expensive thing the cell does âUse information in m. RNA to produce protein âTranslate from nucleic acid to protein âCannot directly template like in replication/transcription

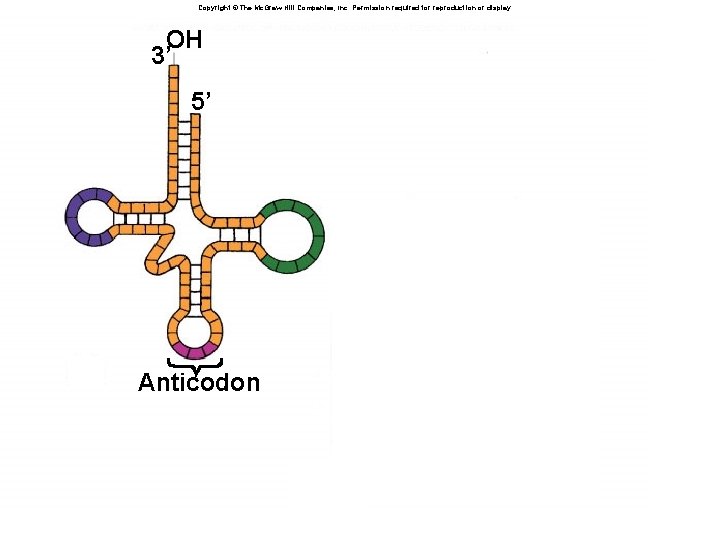
The Essential Adaptor â Nucleic acids cannot directly template aa â Must have molecule to interact with m. RNA and with aa (“read” both languages) â t. RNA is a bifunctional molecule: â One end binds m. RNA: anticodon âBase-pairs with codon in m. RNA â One end binds amino acid: acceptor stem âcovalent bond with carboxyl group of aa â Highly conserved structure â Aminoacyl-t. RNA synthetase â Joins correct amino acid to correct t. RNA â Call the product a charged t. RNA

Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. OH 3’ 5’ 5’ 3’ Anticodon

The Charging Reaction âHow many t. RNA’s do we need to utilize the entire genetic code? âMight think 61… but âActually less because of “wobble rules” âBase pairing in 3 rd position of codon less stringent than other 2 positions âHow many aminoacyl-t. RNA synthetases? âOne for each amino acid âReaction requires energy: ATP

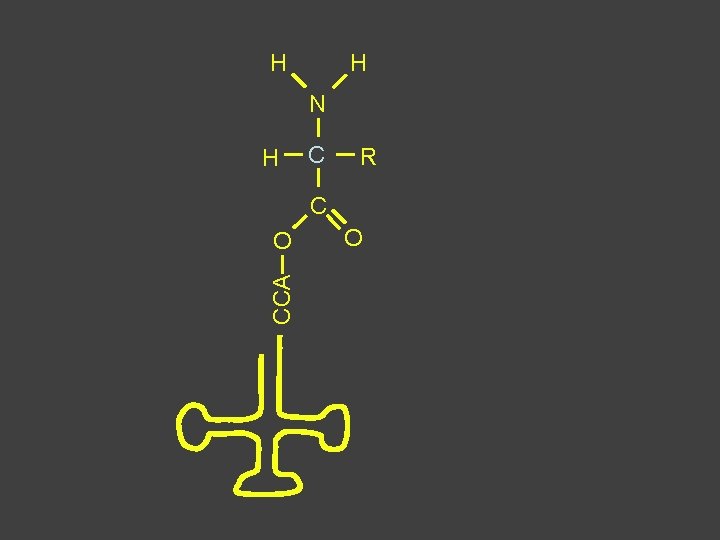
H H N H C R C CCA O O

The Ribosome âThe factory floor for protein synthesis âHuge macromolecular structure composed of: âr. RNA âProteins âOrganized into large and small subunits âCome together during initiation

Ribosome Function âTwo functions: decoding, and enzymatic âMust assemble and put into proper spatial arrangement: âm. RNA (small subunit) â 2 charged t. RNAs (both subunits) âPeptidyl transferase activity (large subunit)

Ribosome Binding Sites ât. RNA binding sites: need at least two, have at least 3 âP site (peptidyl): binds t. RNA with the growing chain (or the first aa) prior to peptide bond âA site (aminoacyl): binds t. RNA with next aa to be added prior to making a peptide bond âE site: this is where the empty t. RNA will exit the ribosome after peptide bond

Copyright © The Mc. Graw-Hill Companies, Inc. Permission required for reproduction or display. E 5’ NH 2 P A 3’ COOH

Phases of Protein Synthesis âInitiation: assembly of the ribosome/m. RNA/charged-t. RNA complex âElongation: formation of successive peptide bonds (add amino acids) âTermination: disassemble complex, cleave peptide from last t. RNA

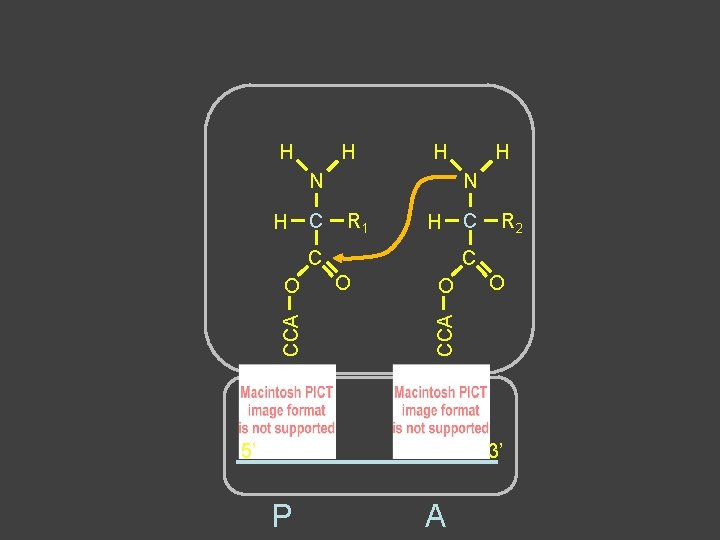
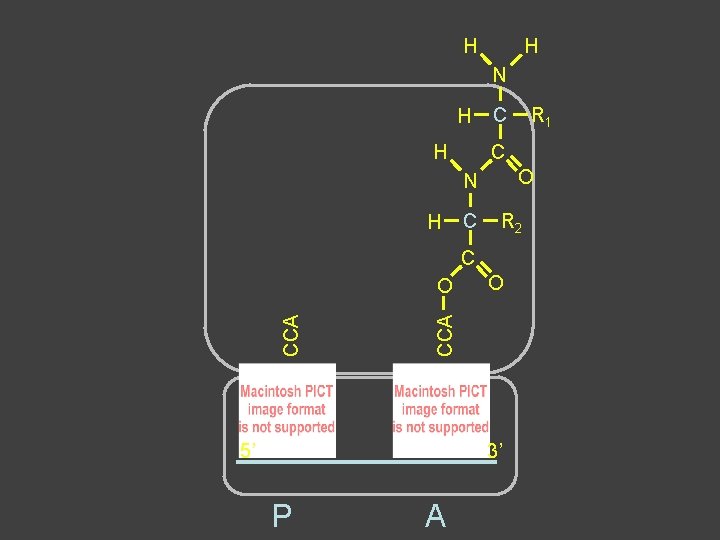
Peptide Bond Formation âAmino terminus of 2 nd aa forms covalent bond with carboxyl terminus of 1 st aa â 2 nd aa remains attached to t. RNA âBond between growing chain and t. RNA is broken in the process âFirst t. RNA-aa (growing chain) is in P site âIncoming t. RNA-aa is in A site âGrowing chain is “transferred” to A site ât. RNA in P site left “empty”

H H H N H C N R 1 H C C R 2 C O O O CCA O H 5’ 3’ P A

H H N H H C C O N H C R 2 C O CCA O 5’ 3’ P A R 1

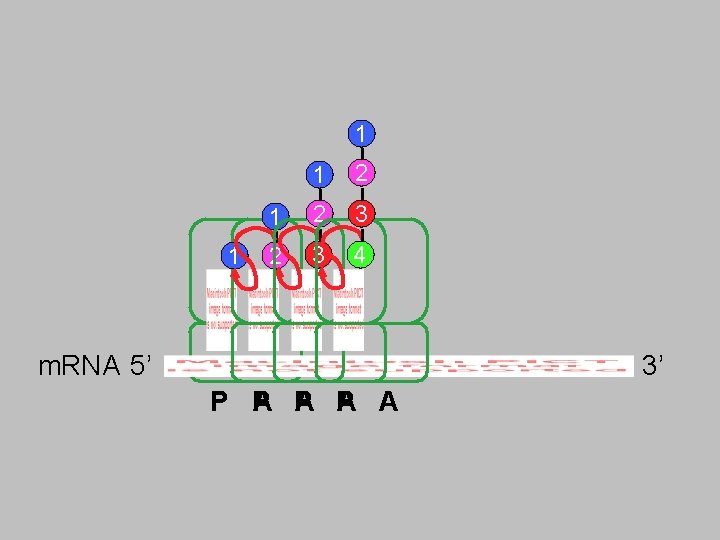
Elongation âStart with growing chain attached to t. RNA in P site 1. Bring new charged t. RNA into A site âMust match codon/anticodon 2. Form peptide bond âGrowing chain now attached to t. RNA in A site ât. RNA in P site now has no aa (empty) 3. Move ribosome relative to m. RNA âBrings new codon into A site âEject empty t. RNA

1 1 1 2 3 2 3 4 m. RNA 5’ 3’ A P A A P P A P
