DNA structure Tertiary Structure topology of plasmids Properties












- Slides: 25

DNA structure: Tertiary Structure: topology of plasmids Properties of Agarose Lecture 10: 1 10/18/2006 Methylation of agarose The agarose polysaccharide also contains uncharged methyl groups. The extent of natural methylation is directly proportional to the gelling temperature. Unexpectedly, synthetically methylated agaroses have lower, rather than higher, gelling temperatures, and the degree of synthetic methylation is inversely proportional to the gelling temperature.

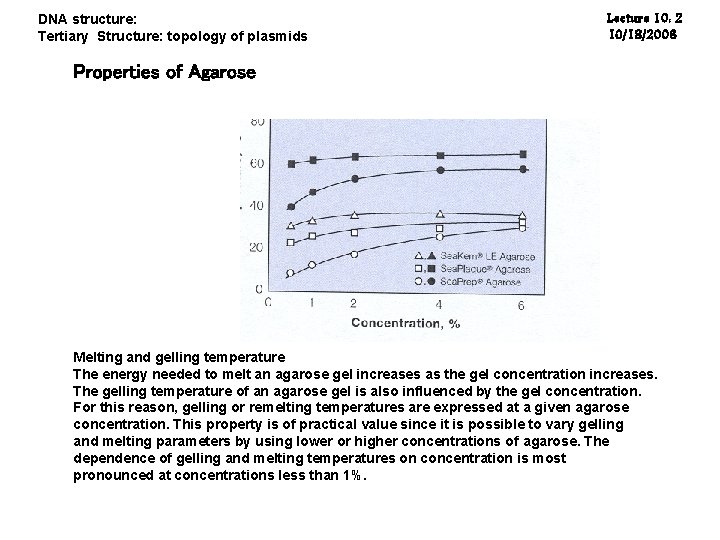
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 2 10/18/2006 Properties of Agarose Melting and gelling temperature The energy needed to melt an agarose gel increases as the gel concentration increases. The gelling temperature of an agarose gel is also influenced by the gel concentration. For this reason, gelling or remelting temperatures are expressed at a given agarose concentration. This property is of practical value since it is possible to vary gelling and melting parameters by using lower or higher concentrations of agarose. The dependence of gelling and melting temperatures on concentration is most pronounced at concentrations less than 1%.

Lecture 10: 2 10/18/2006 DNA structure: Tertiary Structure: topology of plasmids Properties of Agarose Advantages of agarose • Agarose forms a macroporous matrix which allows rapid diffusion of high molecular weight (106 dalton range) macromolecules without significant restriction by the gel. • Agarose gels have a high gel strength, allowing the use of concentrations of 1% or less, while retaining sieving and anticonvective properties. • Agarose is nontoxic and, unlike polyacrylamide, contains no potentially damaging polymerization by-products. There is no free radical polymerization involved in agarose gelation. • Rapid staining and destaining can be performed with minimal background. • Agarose gels are thermoreversible. Low gelling and melting temperature agaroses permit easy recovery of samples, including sensitive heat-labile materials. • Agarose gels may be air dried.

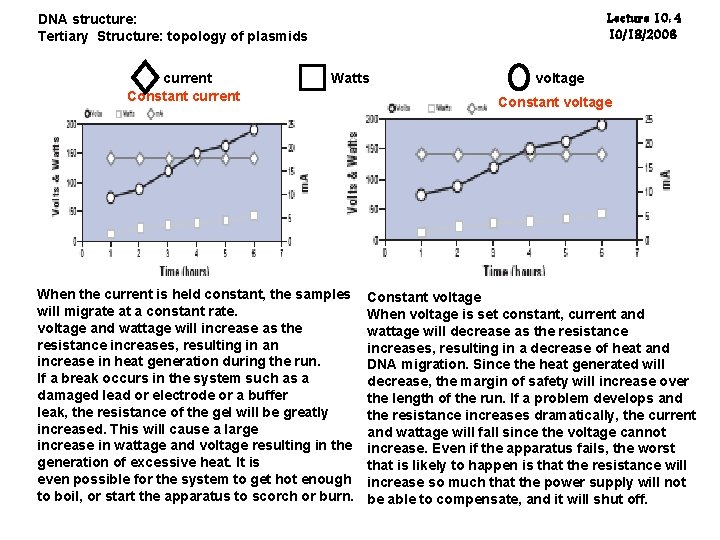
Lecture 10: 4 10/18/2006 DNA structure: Tertiary Structure: topology of plasmids current Constant current Watts When the current is held constant, the samples will migrate at a constant rate. voltage and wattage will increase as the resistance increases, resulting in an increase in heat generation during the run. If a break occurs in the system such as a damaged lead or electrode or a buffer leak, the resistance of the gel will be greatly increased. This will cause a large increase in wattage and voltage resulting in the generation of excessive heat. It is even possible for the system to get hot enough to boil, or start the apparatus to scorch or burn. voltage Constant voltage When voltage is set constant, current and wattage will decrease as the resistance increases, resulting in a decrease of heat and DNA migration. Since the heat generated will decrease, the margin of safety will increase over the length of the run. If a problem develops and the resistance increases dramatically, the current and wattage will fall since the voltage cannot increase. Even if the apparatus fails, the worst that is likely to happen is that the resistance will increase so much that the power supply will not be able to compensate, and it will shut off.

Lecture 10: 5 10/18/2006 DNA structure: Tertiary Structure: topology of plasmids Agarase β-Agarase I digests agarose, releasing trapped DNA and producing carbohydrate molecules which can no longer gel. β-Agarase I can be used to purify both large (> 50 kb) and small (< 50 kb) fragments of DNA from gels. The remaining carbohydrate molecules and βAgarase I will not, in general, interfere with subsequent DNA manipulations such as restriction endonuclease digestion, ligation, and transformation. Q 2: What is the molecular weight of β-Agarase I? A 2: 30, 000 Daltons Q 3: What type of agarose will β-Agarase I digest? A 3: Only low melting point agarose is suitable for β-Agarase I digestion as the solution must be liquid at the incubation temperature of 42°C. If the temperature falls below 42°C during the reaction time, even low melting point agarose will begin to congeal and be undigestable.

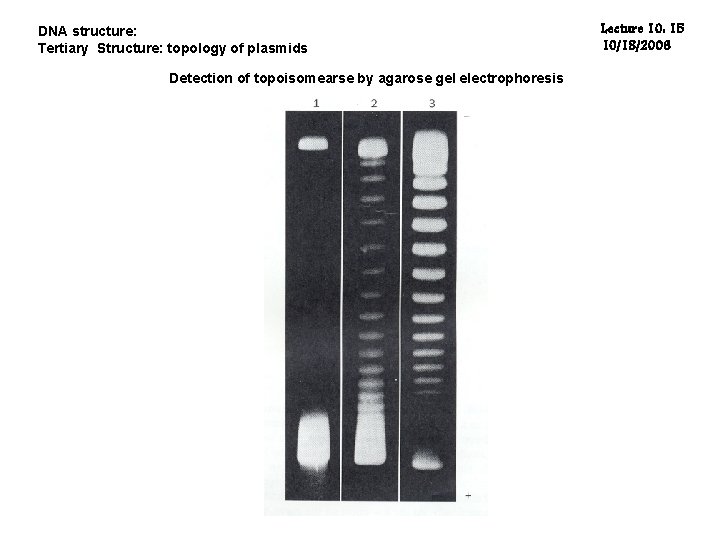
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 6 10/18/2006 Five Possible Conformations are listed below in order of electrophoretic mobility (speed for a given applied voltage) from slowest to fastest: 1. "Nicked Open-Circular" DNA has one strand cut. 2. "Linear" DNA has free ends, either because both strands have been cut, or because the DNA was linear in vivo. 3. "Relaxed Circular" DNA is fully intact with both strands uncut, but has been enzymatically "relaxed" (supercoils removed). 4. "Supercoiled Denatured" DNA is like supercoiled DNA (see below), but has unpaired regions that make it slightly less compact; this can result from excessive alkalinity during plasmid preparation. 5. * "Supercoiled" (or "Covalently Closed-Circular") DNA is fully intact with both strands uncut, and with a twist built in, resulting in a compact form.

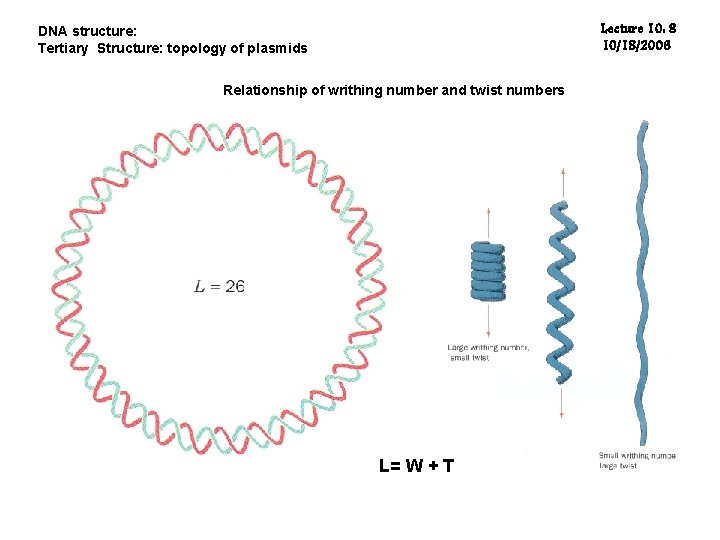
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 7 10/18/2006 Supercoils: a duplex is twisted in space around its own axis. Negative supercoils: the DNA is twisted about its axis in the direction opposite to the intrinsic winding of the helix, thus releasing the torsional pressure or unwinding the helix. Positive supercoils: the DNA is supercoiled in the same direction in The same direction as the intrinsic winding of the double helix. The linking number (L): the number of times that the two strands of the double helix of a closed molecule cross each other in total. L= W + T T: the twisting number, the total number of turns of the duplex W: the writhing number, the turning of the axis of the duplex in space. For relaxed, W = 0. ΔL = ΔW + ΔT, ΔL>0: positive supercoiling; ΔL<0: negative supercoiling Some 10 -50 kcal/mol are needed to separate 10 bp.

Lecture 10: 8 10/18/2006 DNA structure: Tertiary Structure: topology of plasmids Relationship of writhing number and twist numbers L= W + T

DNA structure: Tertiary Structure: topology of plasmids Introducing one supercoil into a DNA with 10 duplex turns Lecture 10: 9 10/18/2006

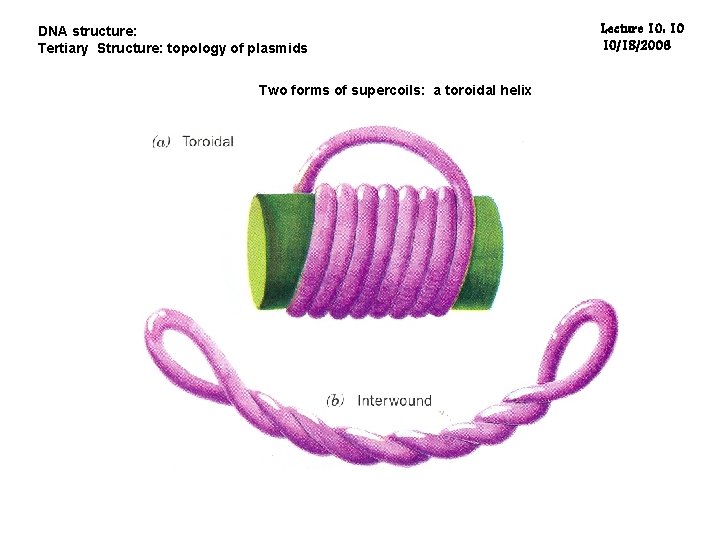
DNA structure: Tertiary Structure: topology of plasmids Two forms of supercoils: a toroidal helix Lecture 10: 10 10/18/2006

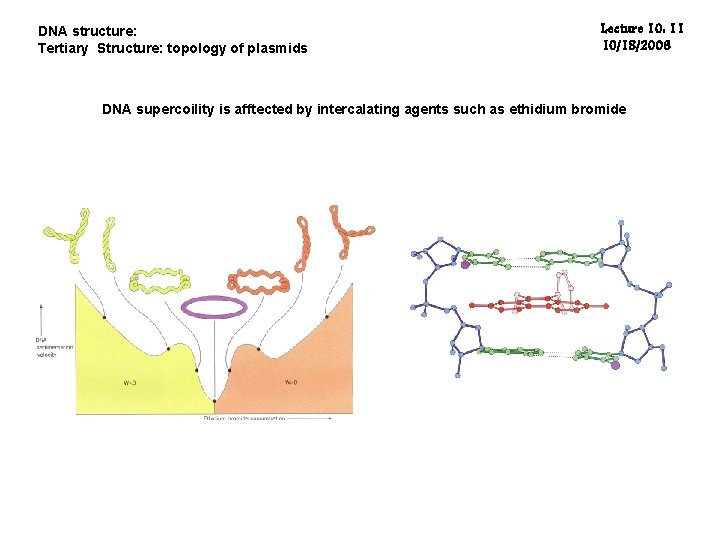
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 11 10/18/2006 DNA supercoility is afftected by intercalating agents such as ethidium bromide

DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 12 10/18/2006 A number of natural and essential cellular processes can have adverse effects on DNA superstructure. 1) the double-helical structure of DNA contorts and supercoils when unwound by polymerases or helicases. 2) Replication of the two complementary DNA strands, or recombination between DNA duplexes, can generate chromosomal knots and catenanes. Failure to resolve these problems can promote misregulation of gene expression and chromosomal breakage, and can have severe consequences for cell viability.

JAMES WANG Mallinckrodt Professor of Biochemistry and Molecular Biology Harvard University Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14; 55(3): 523 -33 Discovering Topoisomerase The late 1960 s were an interesting time to work on campus, according to Wang, a professor of chemistry at Berkeley from 19661977, and currently a professor of molecular and cell biology at Harvard. "There were frequent demonstrations on campus, and once the campus was tear-gassed by a helicopter. For weeks a 'stink bomb' left a repugnant smell in our elevator, and one day a band of demonstrators snaked through the chemistry buildings to look for 'military lasers' that never existed, " he recalled in Reflections on an accidental discovery. Wang discovered topoisomerase while at Berkeley in 1971. His finding attracted a lot of attention because scientists now had a way to separate isomers of DNA that differed only topologically. "Because of my physicochemical background, all enzymes seemed rather mysterious and would better be left to others with the right aptitude for messier systems. Therefore, shortly after my arrival in Berkeley, I decided to try a chemical approach of using a reagent carbodiimide for the desired water splitting. I failed miserably in that expedition.

DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 14 10/18/2006 "The finding of topoisomerase was completely accidental. I was studying negative supercoiling of DNA and had one single cell preparation that was not in agreement with the others. In experimental sciences, a common dictum is 'repeat the experiment if the result makes no sense. ' Is a strange observation reproducible to allow rigorous scrutiny by the methods of science? "When I went back to my notebook to see what was different about this one sample, I noticed that I had spun the cell lysate in the centrifuge for much longer than usual and at a higher temperature. The longer time was due to the fact that I suddenly had to take my daughter to the hospital and had set the machine on 'hold'. The increased temperature was most likely due to the fact that I didn't set the temperature correctly-centrifuges were very bulky and demanding back then-as I rushed off to the hospital. " This single sample lacked negative supercoiling, and it was reasonable for Wang to assume that an activity in the cell, at this incubation, was capable of removing the supercoiling. "After months with the chromatography columns in the cold room, I isolated the active fraction and found that it could indeed relax DNA. "The manuscript was held up for quite some time as the journal's reviewers took their time to 'believe' the results. I can imagine their bewilderment with this strange report of an unprecedented enzyme by someone with no track record in enzymology. For one whole year, I would wonder whether one day the whole thing would turn out to be an artifact. After one year, however, I myself was fully convinced. It probably took many others a few more years to accept the findings, " Wang said.

DNA structure: Tertiary Structure: topology of plasmids Detection of topoisomearse by agarose gel electrophoresis Lecture 10: 15 10/18/2006

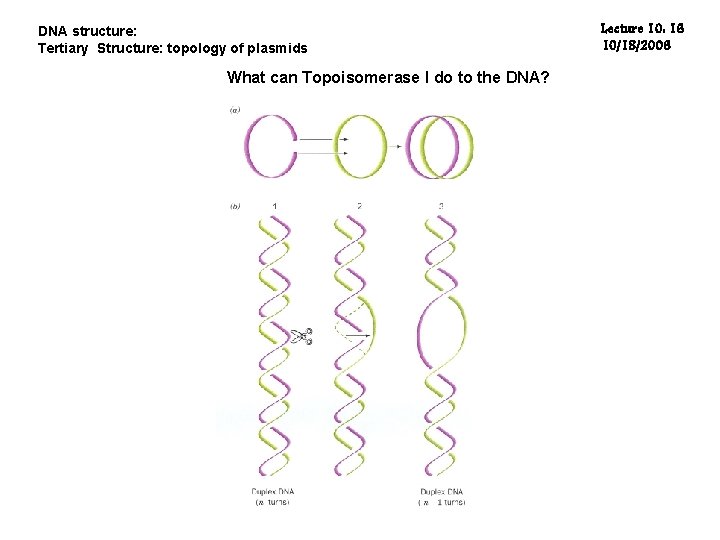
DNA structure: Tertiary Structure: topology of plasmids What can Topoisomerase I do to the DNA? Lecture 10: 16 10/18/2006

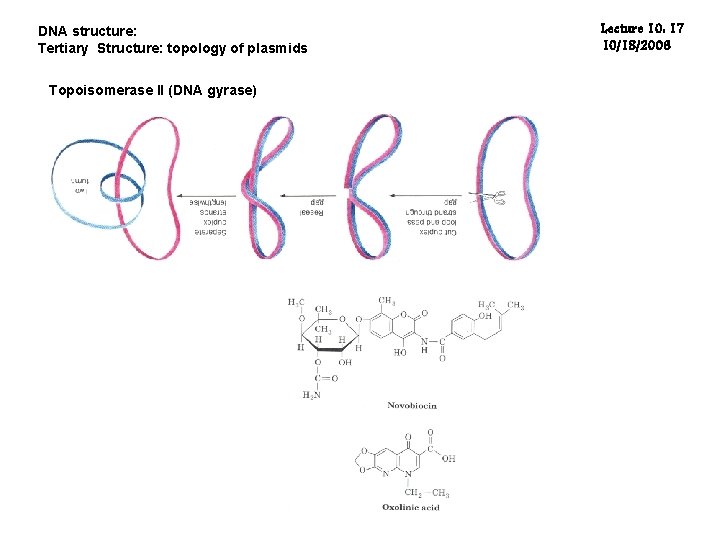
DNA structure: Tertiary Structure: topology of plasmids Topoisomerase II (DNA gyrase) Lecture 10: 17 10/18/2006

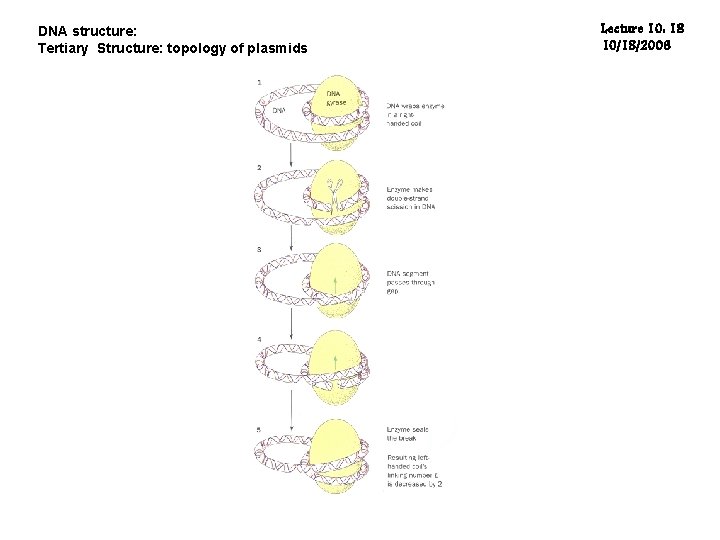
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 18 10/18/2006

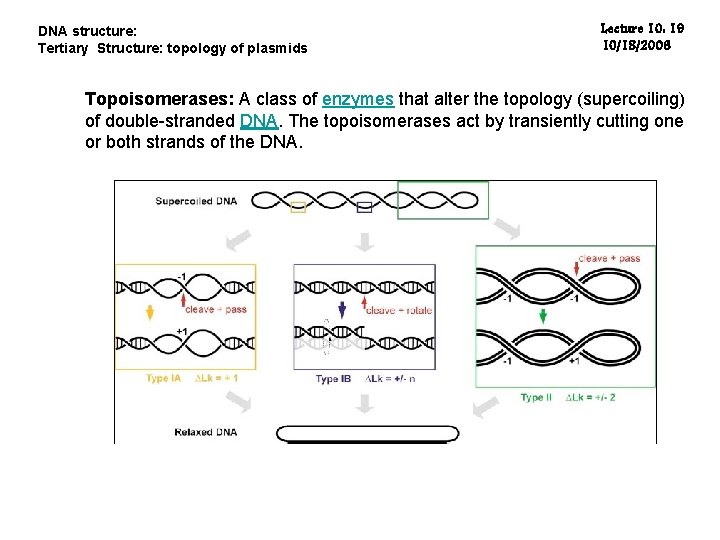
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 19 10/18/2006 Topoisomerases: A class of enzymes that alter the topology (supercoiling) of double-stranded DNA. The topoisomerases act by transiently cutting one or both strands of the DNA.

DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 20 10/18/2006 The intact holoenzyme is a 97 K protein with three Zn(II) atoms in tertacysteine motifs near its carboxy-terminus. Topoisomerase I appears to reverse supercoiling by transiently breaking a segment of single-stranded DNA, passing an intact single- or double-stranded strand of DNA through the gate, then rejoining the broken segment. This is not an active process in the sense that energy in the form of ATP is not spent by the topoisomerase during uncoiling of the DNA; rather, the torque present in the DNA drives the uncoiling. Type I enzymes can be further subdivided into type IA and type IB, based on their chemistry of action. Type IA topoisomerases change the linking number of a circular DNA strand by units of strictly 1, wherease Type IB topoisomerases change the linking number by multiples of 1. Type II topoisomerases cut both strands of the DNA helix simultaneously. Once cut, the ends of the DNA are separated, and a second DNA duplex is passed through the break. Following passage, the cut DNA is resealed. This reaction allows type II topoisomerases to increase or decrease the linking number of a DNA loop by 2 units, and promotes chromosome disentanglement.

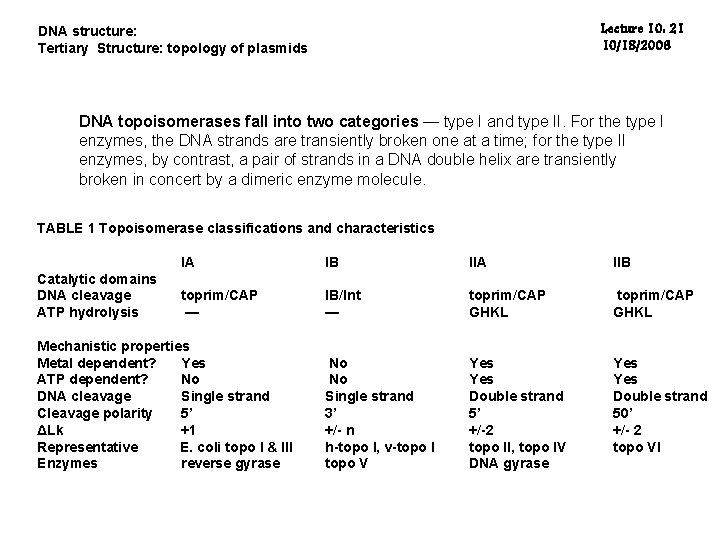
Lecture 10: 21 10/18/2006 DNA structure: Tertiary Structure: topology of plasmids DNA topoisomerases fall into two categories — type I and type II. For the type I enzymes, the DNA strands are transiently broken one at a time; for the type II enzymes, by contrast, a pair of strands in a DNA double helix are transiently broken in concert by a dimeric enzyme molecule. TABLE 1 Topoisomerase classifications and characteristics Catalytic domains DNA cleavage ATP hydrolysis IA IB IIA IIB toprim/CAP — IB/Int — toprim/CAP GHKL No No Single strand 3’ +/- n h-topo I, v-topo I topo V Yes Double strand 5’ +/-2 topo II, topo IV DNA gyrase Yes Double strand 50’ +/- 2 topo VI Mechanistic properties Metal dependent? Yes ATP dependent? No DNA cleavage Single strand Cleavage polarity 5’ ΔLk +1 Representative E. coli topo I & III Enzymes reverse gyrase

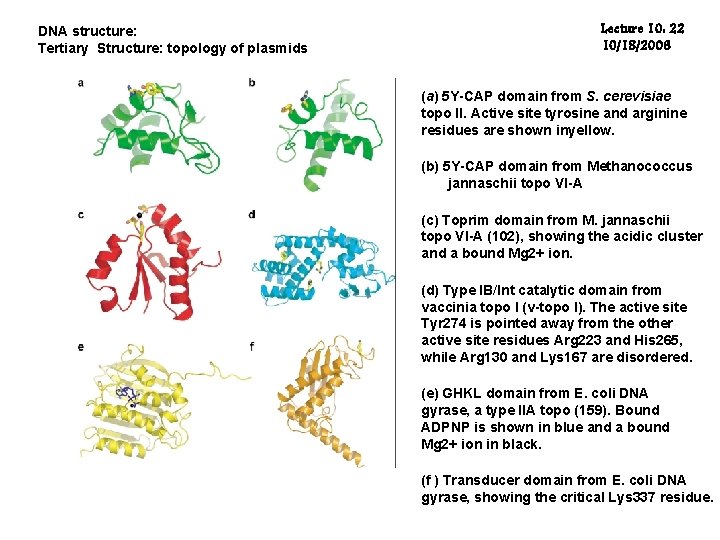
DNA structure: Tertiary Structure: topology of plasmids Lecture 10: 22 10/18/2006 (a) 5 Y-CAP domain from S. cerevisiae topo II. Active site tyrosine and arginine residues are shown inyellow. (b) 5 Y-CAP domain from Methanococcus jannaschii topo VI-A (c) Toprim domain from M. jannaschii topo VI-A (102), showing the acidic cluster and a bound Mg 2+ ion. (d) Type IB/Int catalytic domain from vaccinia topo I (v-topo I). The active site Tyr 274 is pointed away from the other active site residues Arg 223 and His 265, while Arg 130 and Lys 167 are disordered. (e) GHKL domain from E. coli DNA gyrase, a type IIA topo (159). Bound ADPNP is shown in blue and a bound Mg 2+ ion in black. (f ) Transducer domain from E. coli DNA gyrase, showing the critical Lys 337 residue.

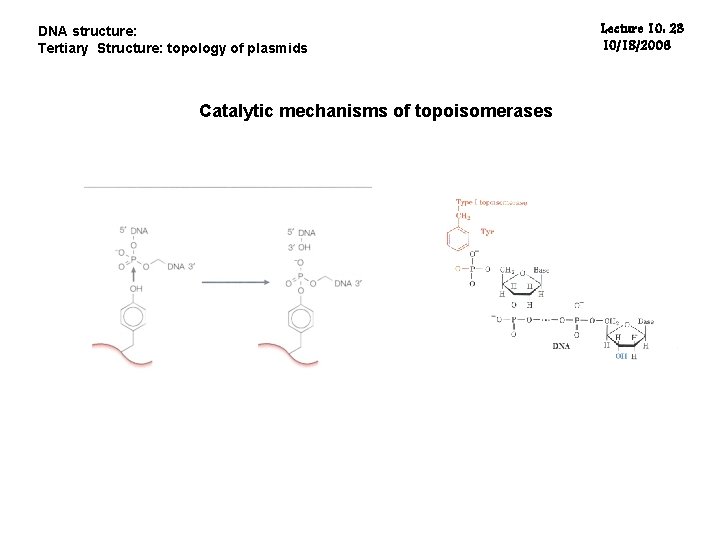
DNA structure: Tertiary Structure: topology of plasmids Catalytic mechanisms of topoisomerases Lecture 10: 23 10/18/2006


 Cosmid vector slideshare
Cosmid vector slideshare Replication fork
Replication fork Bioflix activity dna replication dna replication diagram
Bioflix activity dna replication dna replication diagram Coding dna and non coding dna
Coding dna and non coding dna The principal enzyme involved in dna replication is
The principal enzyme involved in dna replication is Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication Secondary to tertiary structure
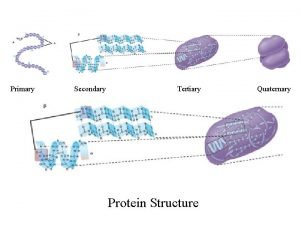
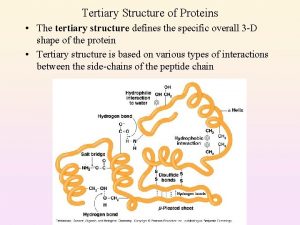
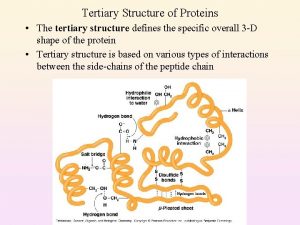
Secondary to tertiary structure Primary vs secondary vs tertiary vs quaternary structures
Primary vs secondary vs tertiary vs quaternary structures Primary secondary and tertiary protein structure
Primary secondary and tertiary protein structure Tertiary storage structure
Tertiary storage structure Protein tertiary structure bonds
Protein tertiary structure bonds Quaternary amine
Quaternary amine Leucine polar or nonpolar
Leucine polar or nonpolar Primary secondary and tertiary structure of protein
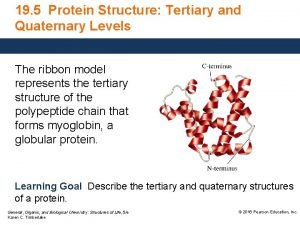
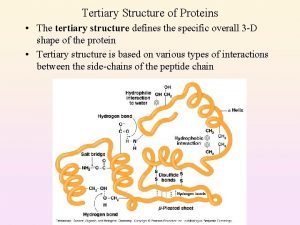
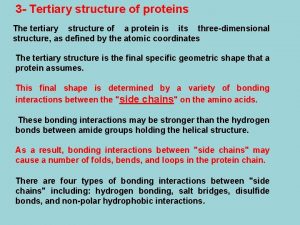
Primary secondary and tertiary structure of protein Tertiary structure
Tertiary structure Tertiary protein structure
Tertiary protein structure Extensive examples
Extensive examples Chemical property of matter
Chemical property of matter Haploid vs diploid
Haploid vs diploid Atp ester bond
Atp ester bond Label dna molecule
Label dna molecule Lego ppt
Lego ppt Describe the structure of dna
Describe the structure of dna Multiple choice questions on dna structure and replication
Multiple choice questions on dna structure and replication Human genome
Human genome Dna structure labeled hydrogen bonds
Dna structure labeled hydrogen bonds