DNA Structure DNA Replication and Repair Genetic Information















- Slides: 50

DNA Structure & DNA Replication and Repair

Genetic Information Genetic information is stored in molecules called nucleic acids. There are 2 types of nucleic acids DNA: deoxyribonucleic acid ◦ Double stranded RNA: ribonucleic acid ◦ Single stranded

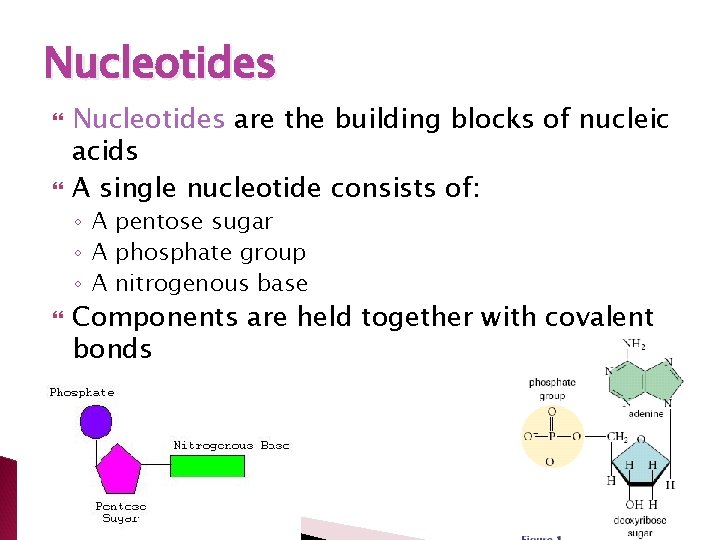
Nucleotides are the building blocks of nucleic acids A single nucleotide consists of: ◦ A pentose sugar ◦ A phosphate group ◦ A nitrogenous base Components are held together with covalent bonds

Nitrogenous Base Also known as an organic base, or a nitrogen base 5 different bases: ◦ ◦ ◦ Adenine (A) Cytosine (C) Guanine (G) Thymine (T) – only found in DNA Uracil (U) – only found in RNA

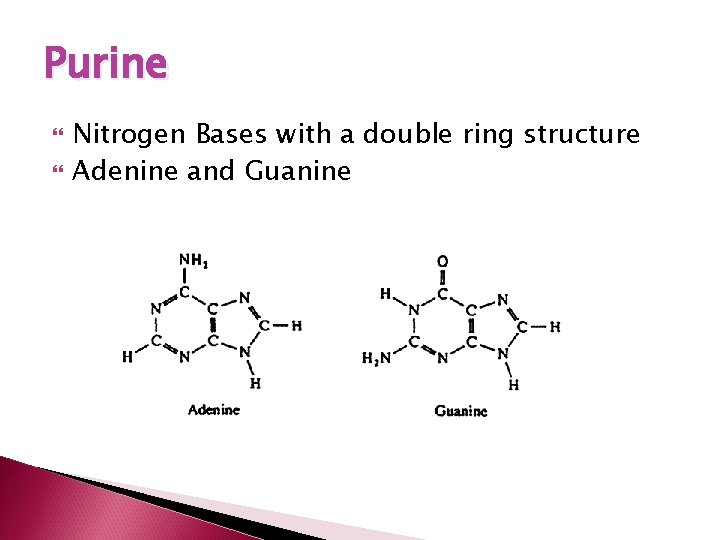
Purine Nitrogen Bases with a double ring structure Adenine and Guanine

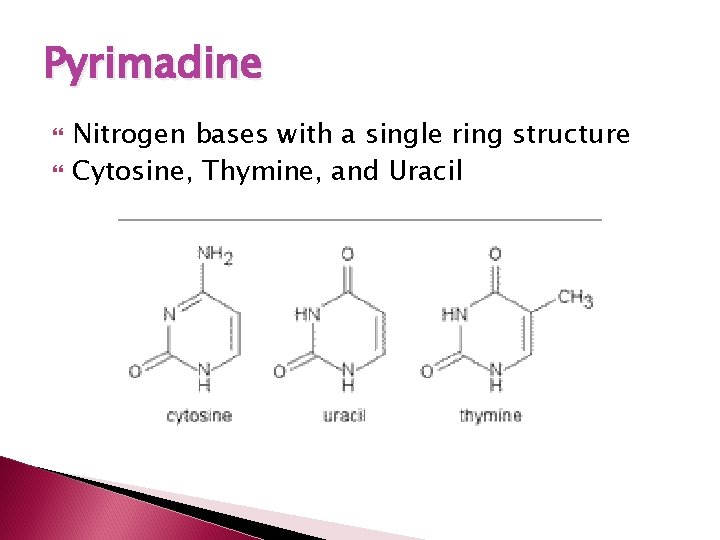
Pyrimadine Nitrogen bases with a single ring structure Cytosine, Thymine, and Uracil

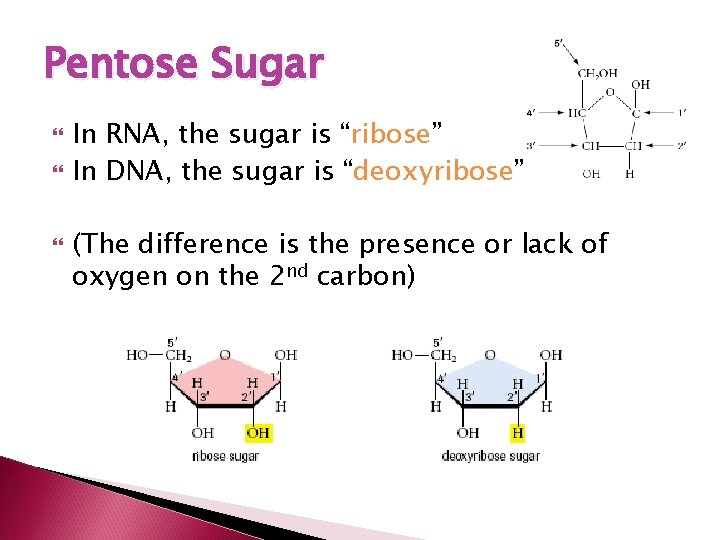
Pentose Sugar In RNA, the sugar is “ribose” In DNA, the sugar is “deoxyribose” (The difference is the presence or lack of oxygen on the 2 nd carbon)

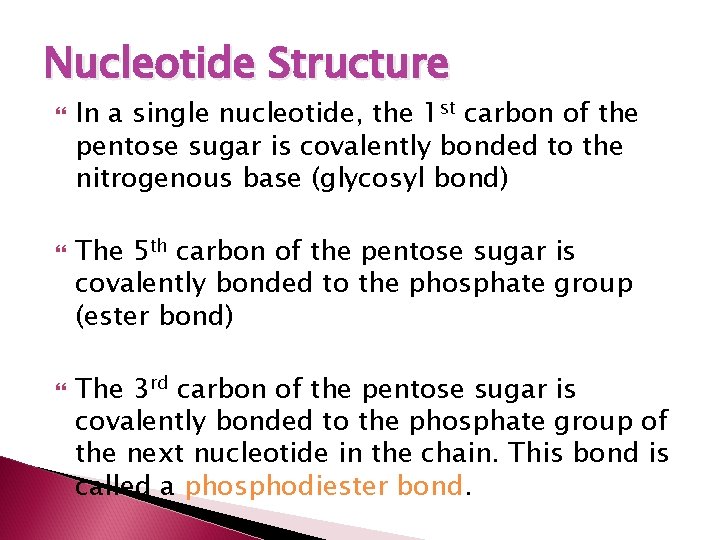
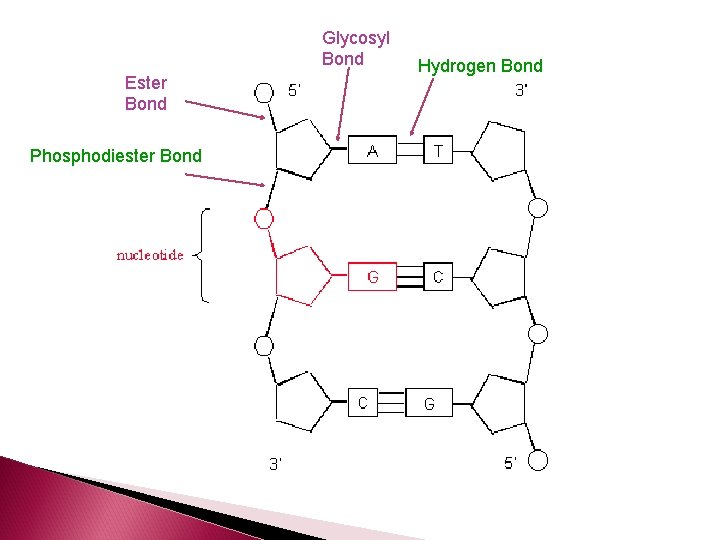
Nucleotide Structure In a single nucleotide, the 1 st carbon of the pentose sugar is covalently bonded to the nitrogenous base (glycosyl bond) The 5 th carbon of the pentose sugar is covalently bonded to the phosphate group (ester bond) The 3 rd carbon of the pentose sugar is covalently bonded to the phosphate group of the next nucleotide in the chain. This bond is called a phosphodiester bond.

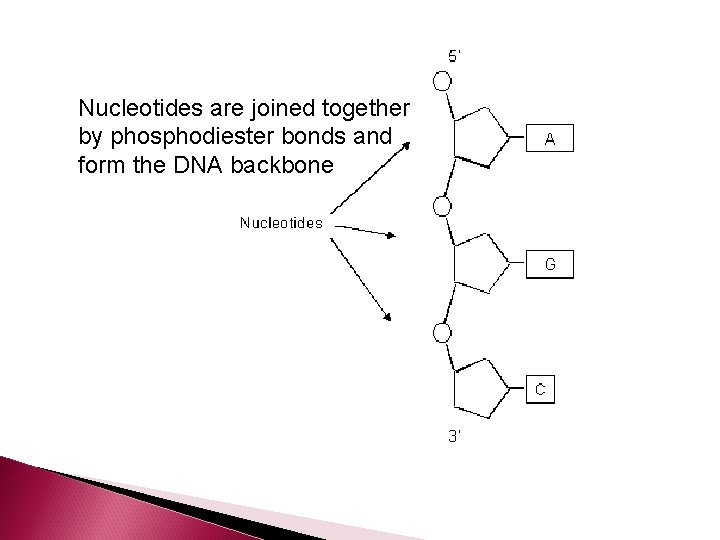
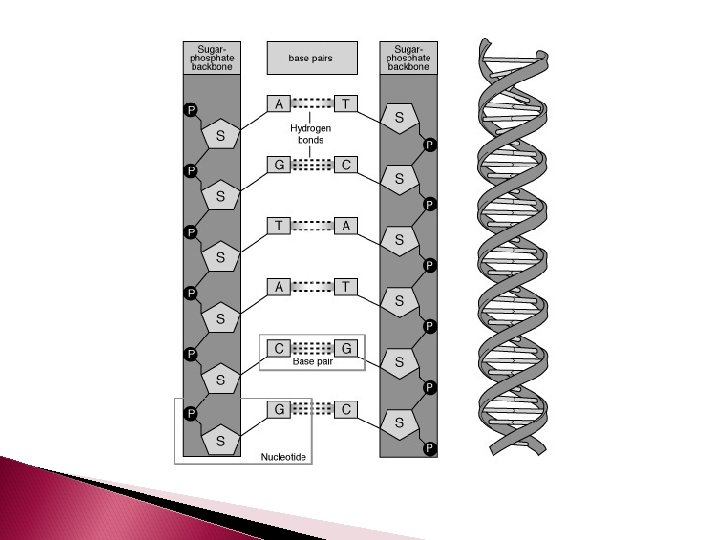
Nucleotides are joined together by phosphodiester bonds and form the DNA backbone

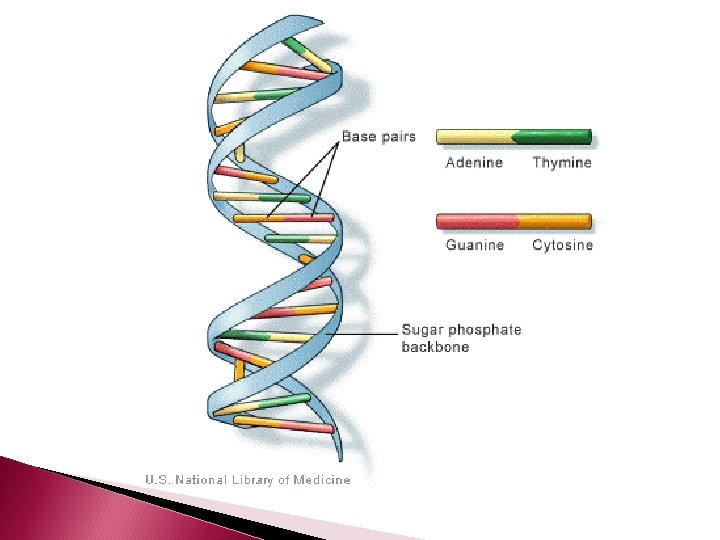
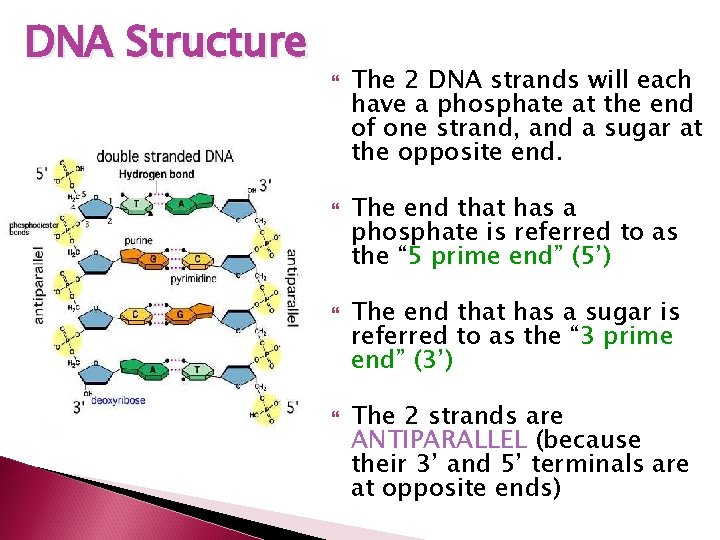
DNA Structure Remember DNA, is double stranded The 2 strands of DNA are complimentary The nitrogen bases of 2 complimentary nucleotides hydrogen bond to each other to create the double strand Adenine always bonds to Thymine and Cytosine always bonds to Guanine

Glycosyl Bond Ester Bond Phosphodiester Bond Hydrogen Bond


DNA Structure There are 2 H bonds between Adenine and Thymine There are 3 H bonds between Guanine and Cytosine How many would you expect uracil to make with a potential nucleotide? DNA forms a double helix The helix is created by H-bonds between nonconsecutive nucleotides


DNA Structure The 2 DNA strands will each have a phosphate at the end of one strand, and a sugar at the opposite end. The end that has a phosphate is referred to as the “ 5 prime end” (5’) The end that has a sugar is referred to as the “ 3 prime end” (3’) The 2 strands are ANTIPARALLEL (because their 3’ and 5’ terminals are at opposite ends)

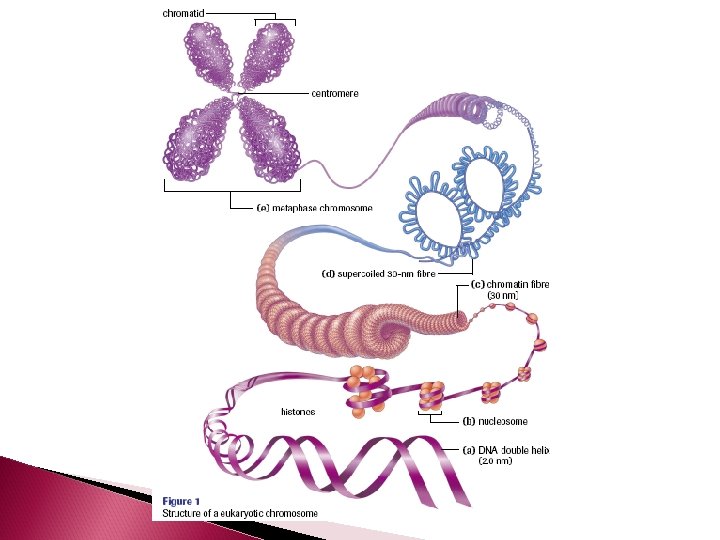
DNA Organization DNA is extremely long If you took the DNA of a single cell and stretched it out into one long double helix, it would measure 1. 8 m in length If fits into a cell because it is very tightly packed – which also keeps it organized!

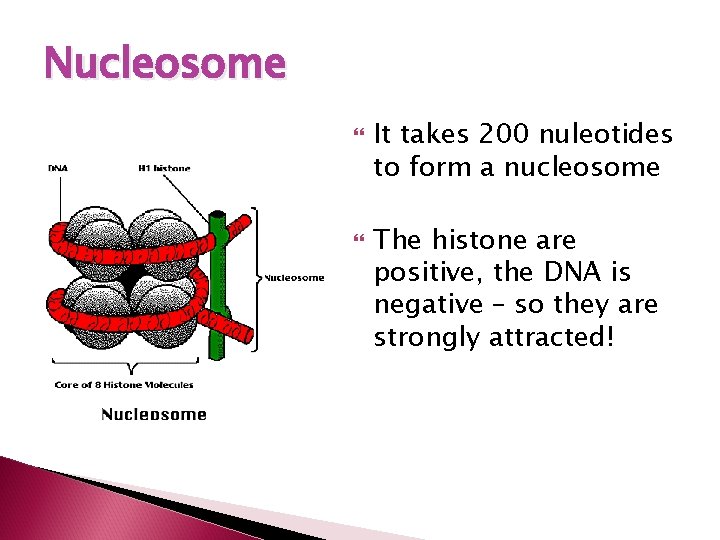
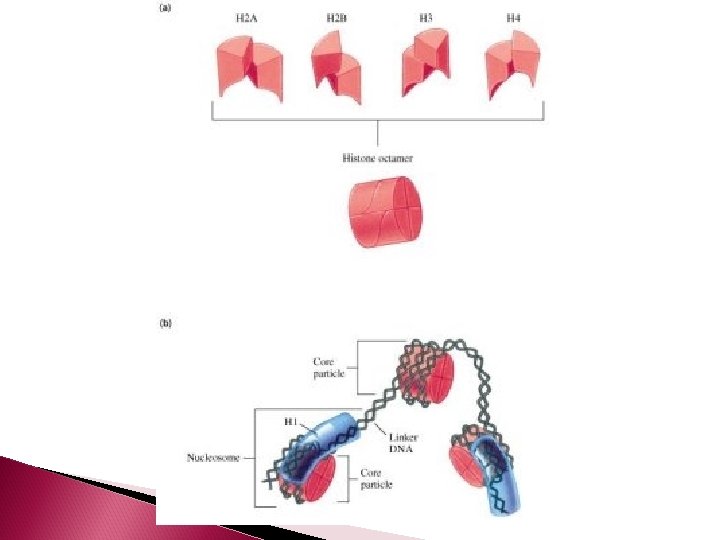
DNA Organization Just like thread is spun around a spool to keep it organized, DNA is coiled around a group of eight proteins called histones. The complex of histones and DNA is called a NUCLEOSOME

Nucleosome It takes 200 nuleotides to form a nucleosome The histone are positive, the DNA is negative – so they are strongly attracted!

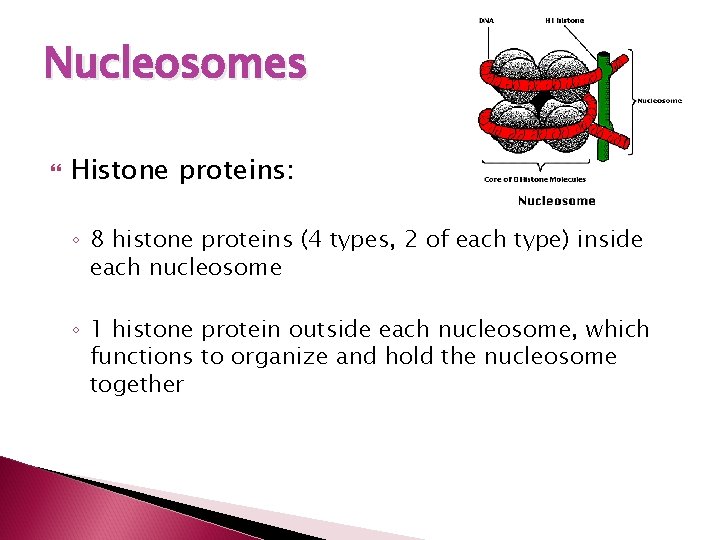
Nucleosomes Histone proteins: ◦ 8 histone proteins (4 types, 2 of each type) inside each nucleosome ◦ 1 histone protein outside each nucleosome, which functions to organize and hold the nucleosome together


DNA Organization A series of nucleosomes coil into chromatin fibres The chromatin fibres then coil to form a supercoil The supercoiled chromatin is what makes up a chromosome A chromosome is one unbroken doublestranded DNA helix


Nucleosome Function Not only do nucleosomes keep DNA organized, they also prevent trancription Transcription is when DNA is used as a template to produce an RNA strand. For this to occur, the enzyme RNA polymerase must attach to the 3’ end of a DNA strand.

When DNA is organized in a nucleosome, the promoter region is inaccessble so transcription cannot take place When the cell requires transcription, enzymes will alter the shape of the nucleosome to allow RNA polymerase to attach.

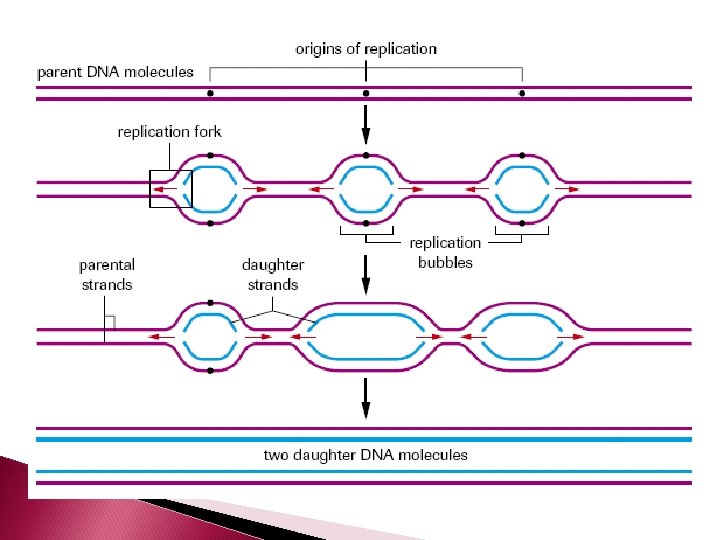
DNA Replication For life to perpetuate, cell must replicate (undergo cell division – mitosis and cytokinesis) Before mitosis can occur, the DNA in the nucleus must duplicate

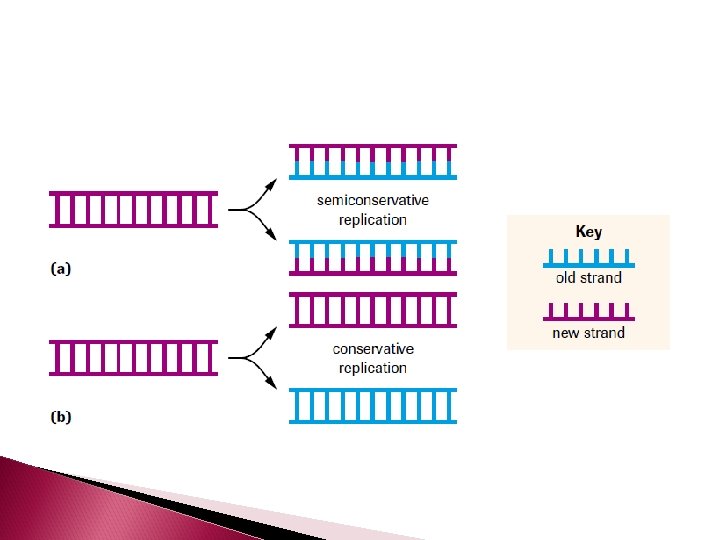
DNA Replcation DNA replication is semiconservative The parent double helix produces 2 daughter double helices. Each daughter molecule will have a parental strand a daughter strand (an old strand a new strand)


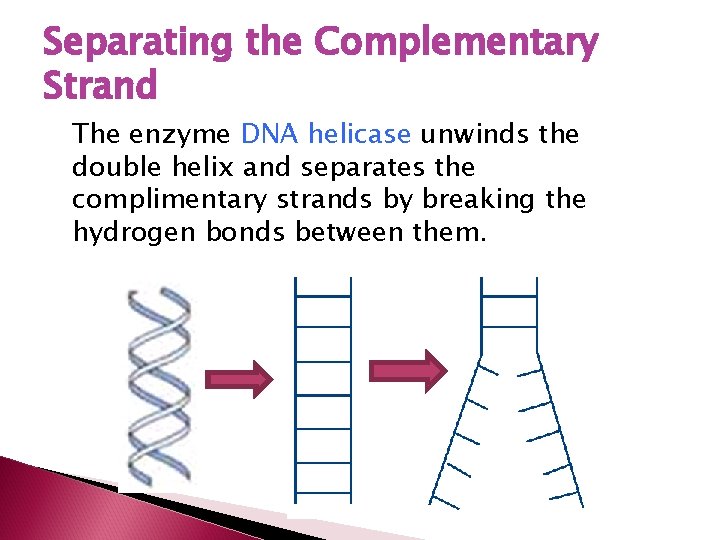
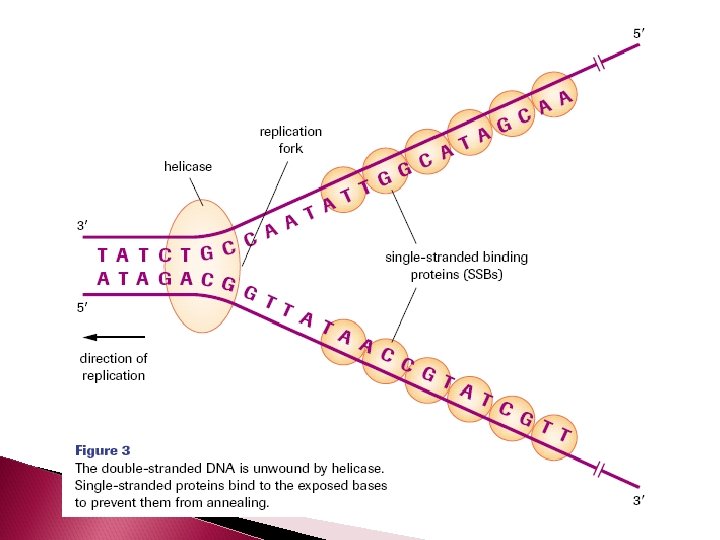
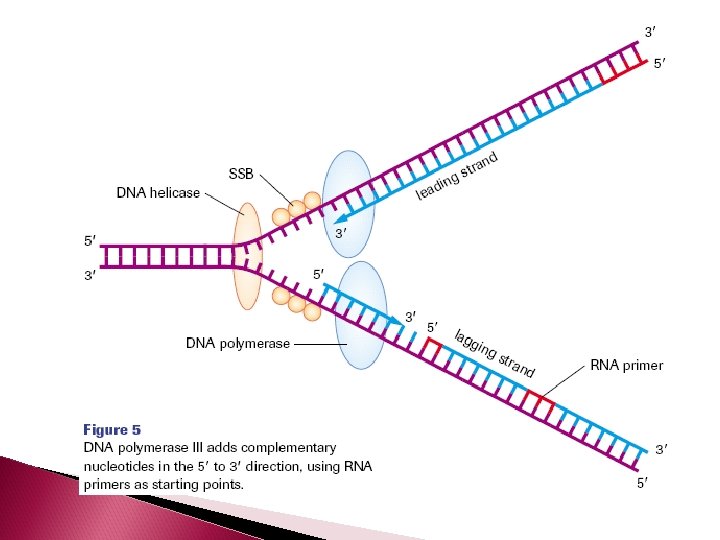
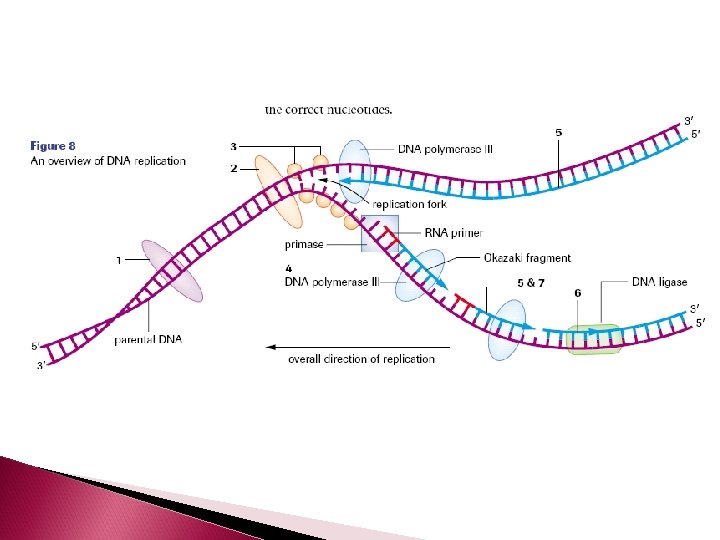
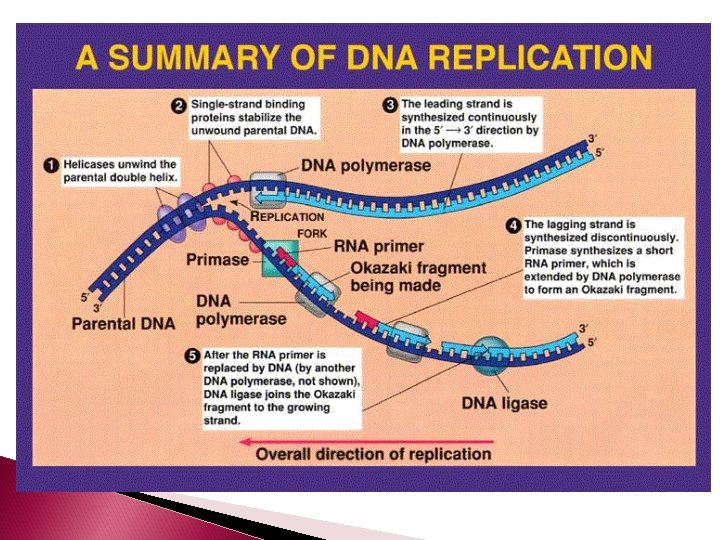
Separating the Complementary Strand The enzyme DNA helicase unwinds the double helix and separates the complimentary strands by breaking the hydrogen bonds between them.

Base pairs have a natural propensity to anneal (stick together) Single-stranded binding protein (SSBs) bind to the exposed DNA single strands and block hydrogen bonding DNA gyrase relieve tension produced by the unwinding of DNA


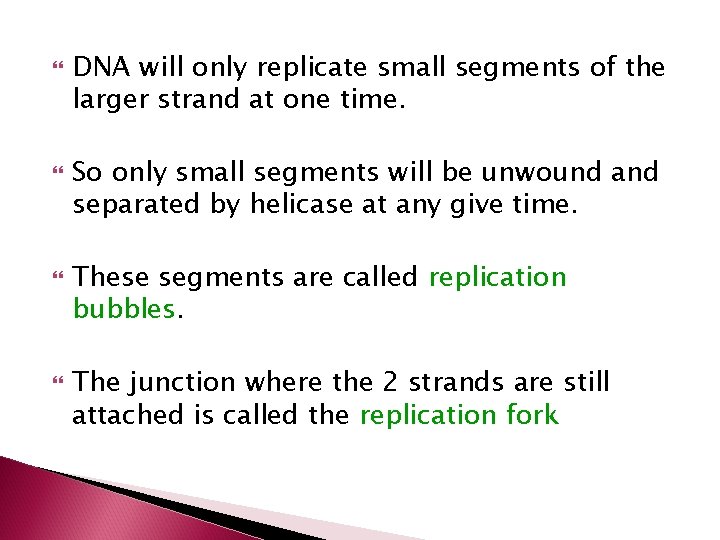
DNA will only replicate small segments of the larger strand at one time. So only small segments will be unwound and separated by helicase at any give time. These segments are called replication bubbles. The junction where the 2 strands are still attached is called the replication fork


Building the Complementary Strands In eukaryotes, five different types of DNA polymerase are present The enzyme that builds the complementary strand using the template strand as a guide is DNA polymerase III

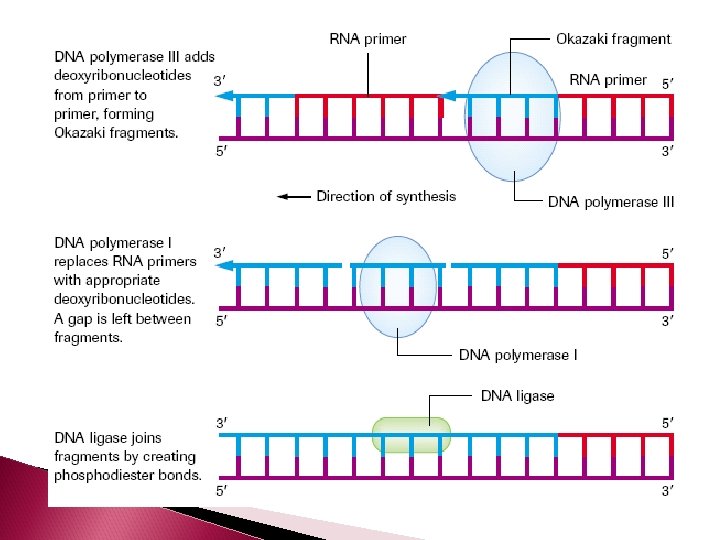
DNA polymerase III cannot initiate a new complementary strand by itself The enzyme primase creates an RNA primer – which is 10 - 60 RNA nucleotides. The RNA primer temporarily attaches to the 3’ end of a DNA strand. The purpose of the primer is to create a starting point for the DNA nucleotides to attach

Once the RNA primer is in place, DNA polymerase III can start elongation by adding free deoxyribonucleotide triphosphates to the growing complementary strand

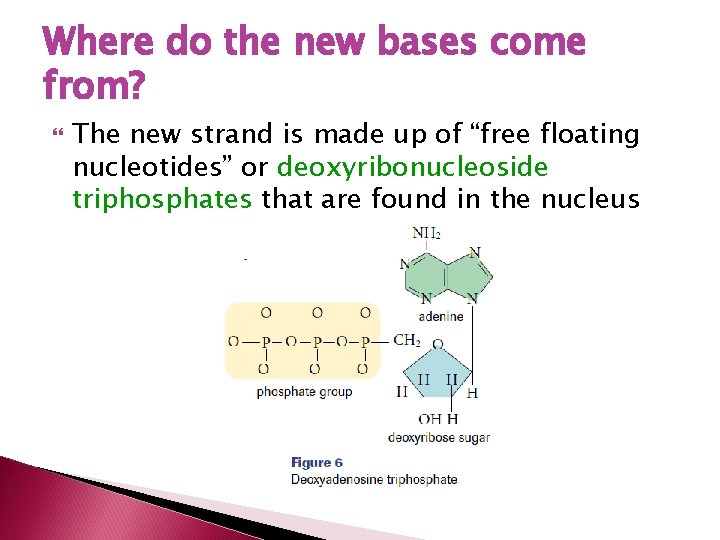
Where do the new bases come from? The new strand is made up of “free floating nucleotides” or deoxyribonucleoside triphosphates that are found in the nucleus

DNA polymerase III uses the energy derived from breaking the bond between the first and second phosphate to drive the condensation reaction that adds a complementary nucleotide to the elongating strand The extra two phosphates are recycled by the cell and are used to build more nucleotide triphosphates

Building the Complementary Strands DNA is always synthesized in the 5’ -3’ direction Since DNA strands run antiparallel, only one strand is able to be built continuously The strand which is able to be continuously synthesized in the 5’ to 3’ direction is called the leading strand


LAGGING STRAND The other strand is called the lagging strand. It is synthesized discontinuously in the direction away from the replication fork and in the opposite direction of helicase. As a result, short fragments (1000 -2000 nucleotides in length) are produced called Okazaki fragments (at the beginning of each Okazaki fragment there will be a RNA primer)

The enzyme DNA polymerase I removes the RNA primers from both the leading and lagging strands and replaces them with the appropriate DNA nucleotides.

On the lagging strand, the enzyme DNA ligase will attach the Okazaki fragments of the lagging strand together. As the 2 new double strands of DNA are made, they will automatically twist into a helix.





Proofreading and Repair DNA polymerase I and III “proofread” the newly created strands checking for mistakes. If there is a mistake, the enzymes act as an exonuclease It removes the incorrect nucleotide and replaces it with the correct one.

Animations http: //highered. mcgrawhill. com/sites/0072437316/student_view 0/c hapter 14/animations. html#

http: //www. youtube. com/watch? v=8 k. K 2 zwj. R V 0 M

Application to Cancer Therapy http: //www. youtube. com/watch? v=NAPYa. SQ Pios