DNA Structure and Function The Molecular Basis of
DNA Structure and Function The Molecular Basis of Inheritance
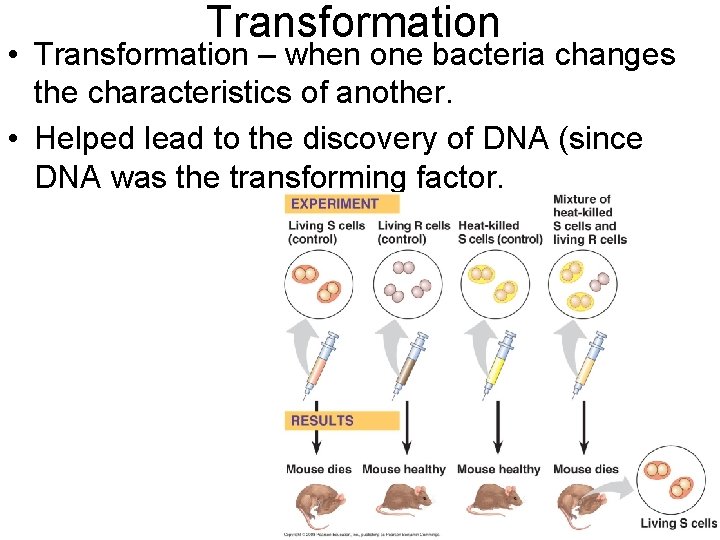
Transformation • Transformation – when one bacteria changes the characteristics of another. • Helped lead to the discovery of DNA (since DNA was the transforming factor.
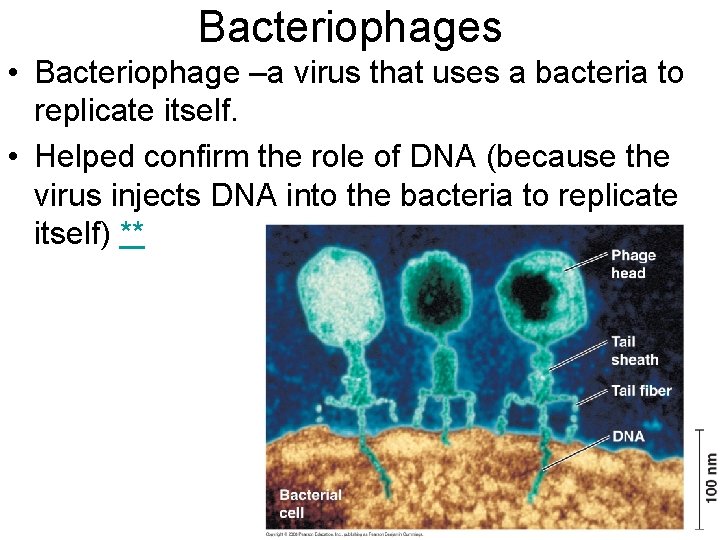
Bacteriophages • Bacteriophage –a virus that uses a bacteria to replicate itself. • Helped confirm the role of DNA (because the virus injects DNA into the bacteria to replicate itself) **
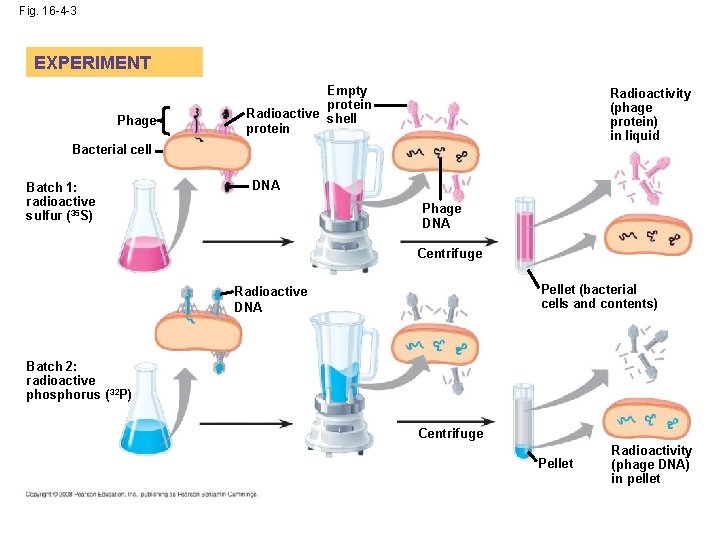
Fig. 16 -4 -3 EXPERIMENT Phage Empty protein Radioactive shell protein Radioactivity (phage protein) in liquid Bacterial cell Batch 1: radioactive sulfur (35 S) DNA Phage DNA Centrifuge Pellet (bacterial cells and contents) Radioactive DNA Batch 2: radioactive phosphorus ( 32 P) Centrifuge Pellet Radioactivity (phage DNA) in pellet
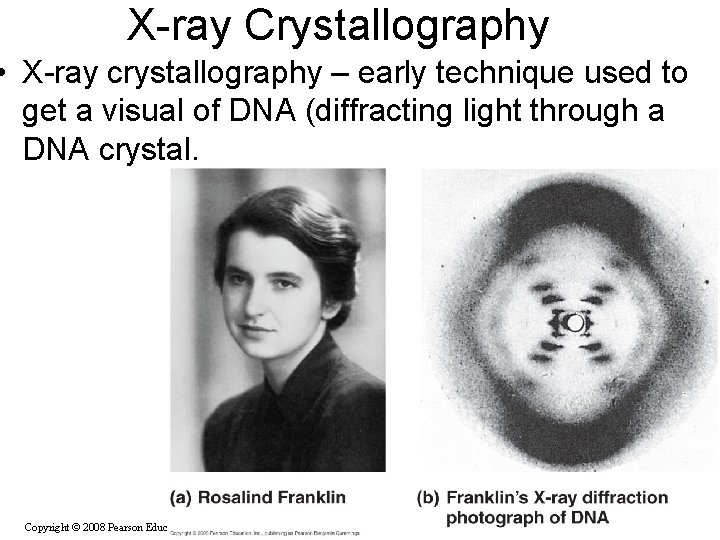
X-ray Crystallography • X-ray crystallography – early technique used to get a visual of DNA (diffracting light through a DNA crystal. Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
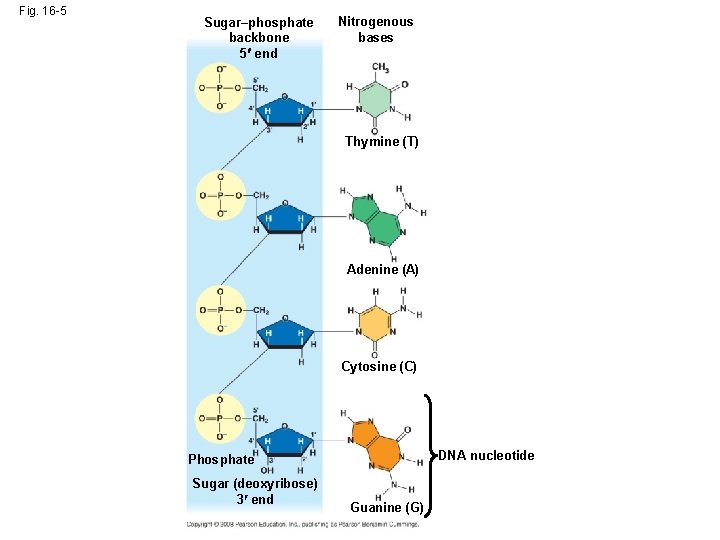
Fig. 16 -5 Sugar–phosphate backbone 5 end Nitrogenous bases Thymine (T) Adenine (A) Cytosine (C) DNA nucleotide Phosphate Sugar (deoxyribose) 3 end Guanine (G)
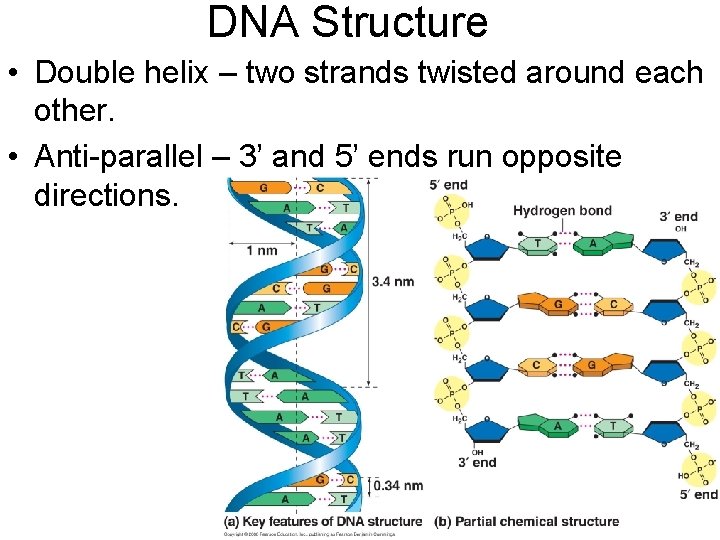
DNA Structure • Double helix – two strands twisted around each other. • Anti-parallel – 3’ and 5’ ends run opposite directions.
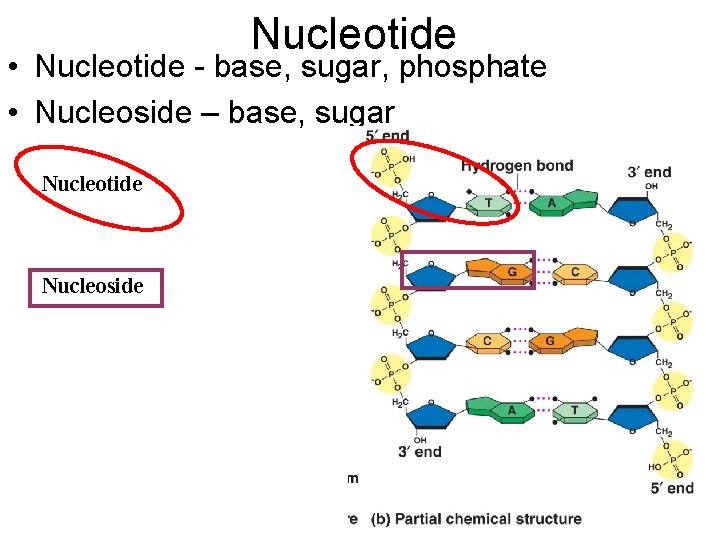
Nucleotide • Nucleotide - base, sugar, phosphate • Nucleoside – base, sugar Nucleotide Nucleoside
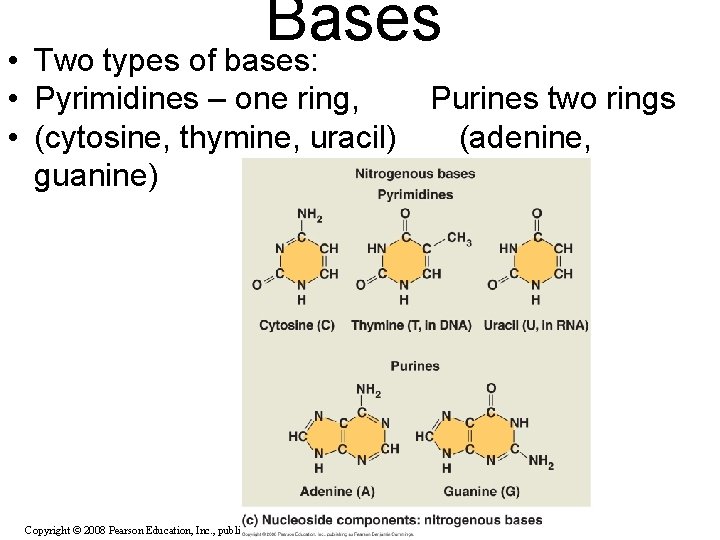
Bases • Two types of bases: • Pyrimidines – one ring, • (cytosine, thymine, uracil) guanine) Copyright © 2008 Pearson Education, Inc. , publishing as Pearson Benjamin Cummings Purines two rings (adenine,
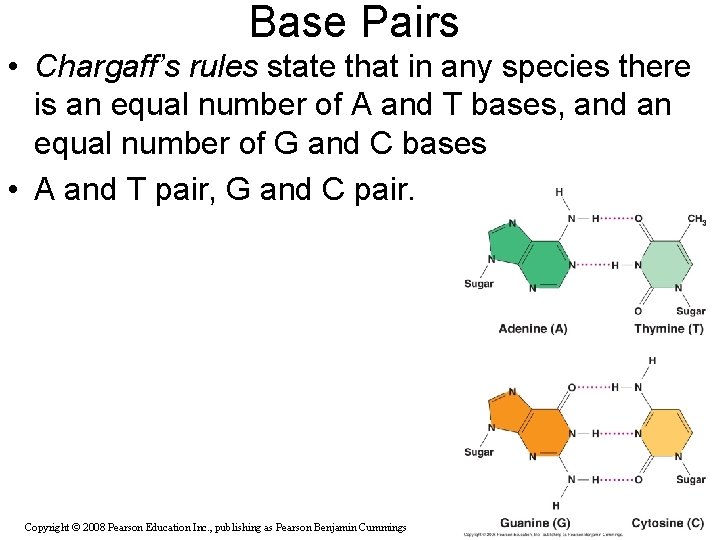
Base Pairs • Chargaff’s rules state that in any species there is an equal number of A and T bases, and an equal number of G and C bases • A and T pair, G and C pair. Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
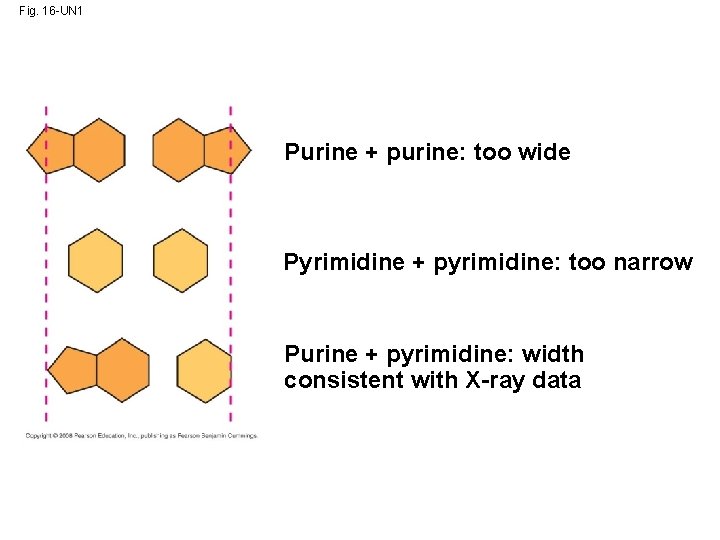
Fig. 16 -UN 1 Purine + purine: too wide Pyrimidine + pyrimidine: too narrow Purine + pyrimidine: width consistent with X-ray data
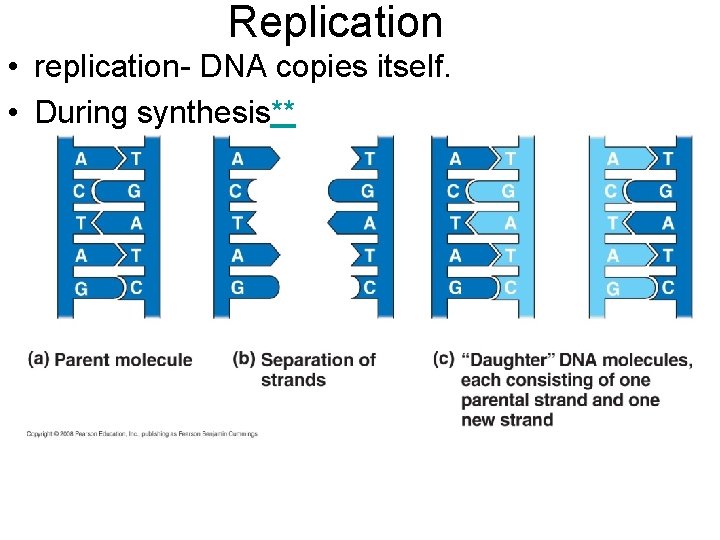
Replication • replication- DNA copies itself. • During synthesis**

Semiconservative Model • semiconservative model – replicated DNA will have one old and one new strand of DNA. (correct model) • conservative model the two parent strands rejoin • dispersive model each strand is a mix of old and new
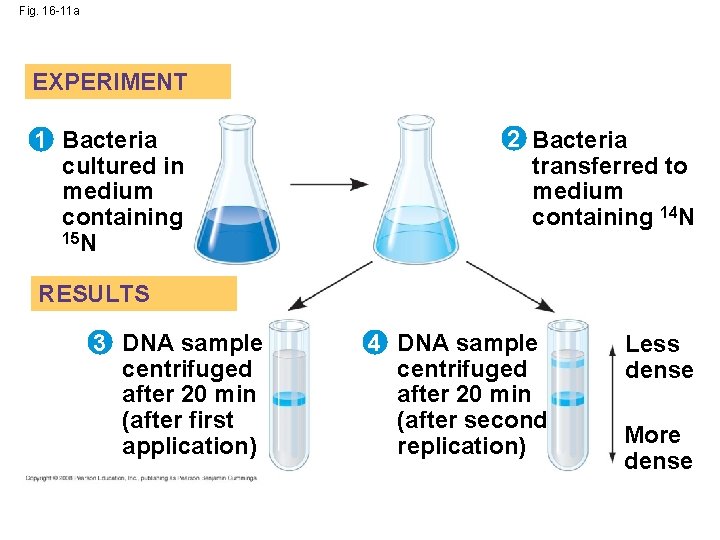
Fig. 16 -11 a EXPERIMENT 1 Bacteria cultured in medium containing 15 N 2 Bacteria transferred to medium containing 14 N RESULTS 3 DNA sample centrifuged after 20 min (after first application) 4 DNA sample centrifuged after 20 min (after second replication) Less dense More dense
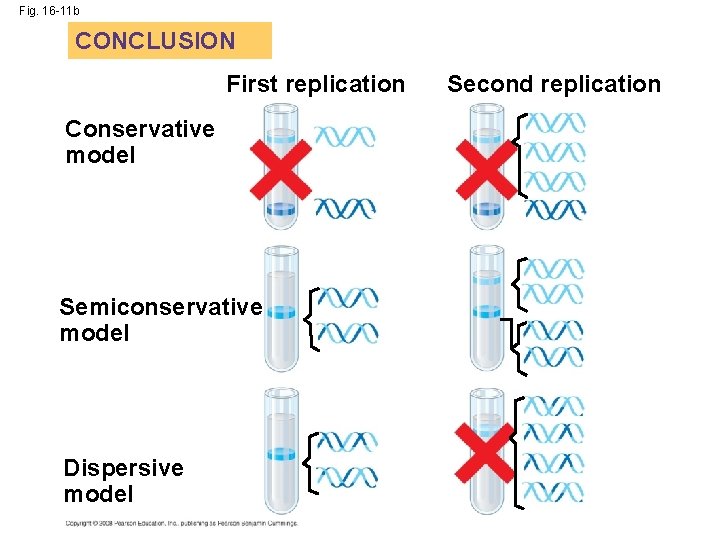
Fig. 16 -11 b CONCLUSION First replication Conservative model Semiconservative model Dispersive model Second replication
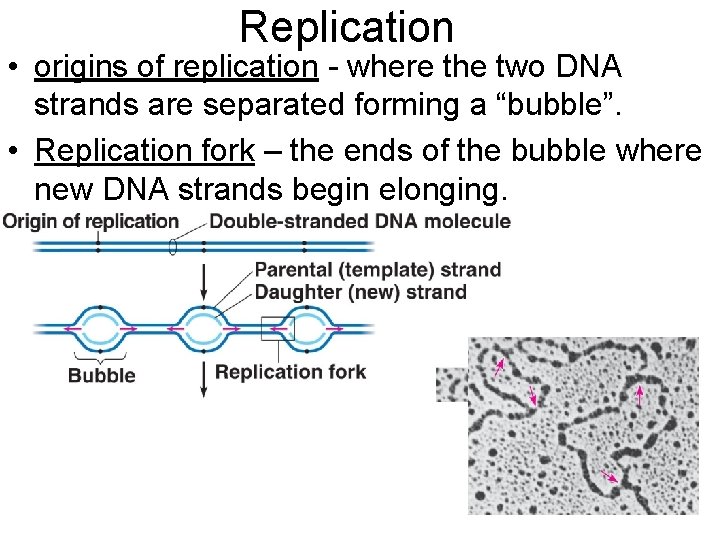
Replication • origins of replication - where the two DNA strands are separated forming a “bubble”. • Replication fork – the ends of the bubble where new DNA strands begin elonging.
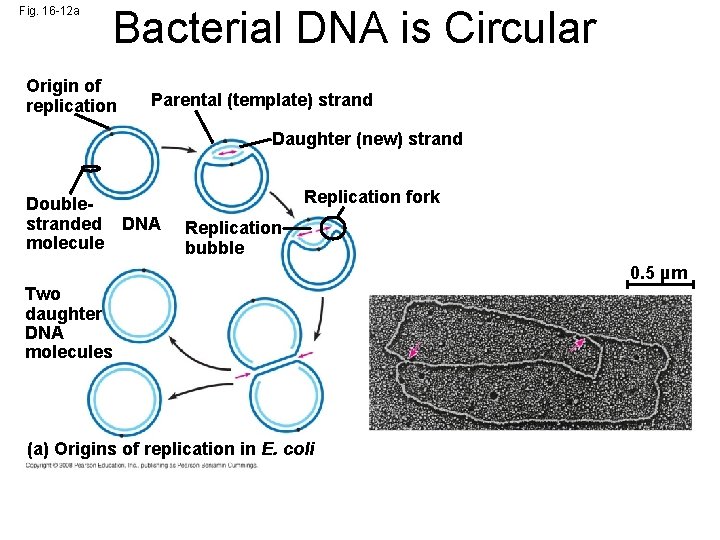
Fig. 16 -12 a Bacterial DNA is Circular Origin of replication Parental (template) strand Daughter (new) strand Doublestranded DNA molecule Replication fork Replication bubble 0. 5 µm Two daughter DNA molecules (a) Origins of replication in E. coli
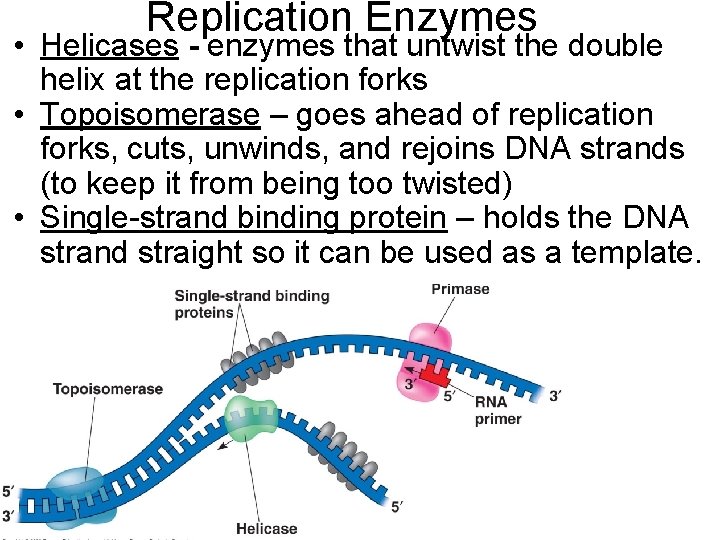
Replication Enzymes • Helicases - enzymes that untwist the double helix at the replication forks • Topoisomerase – goes ahead of replication forks, cuts, unwinds, and rejoins DNA strands (to keep it from being too twisted) • Single-strand binding protein – holds the DNA strand straight so it can be used as a template.
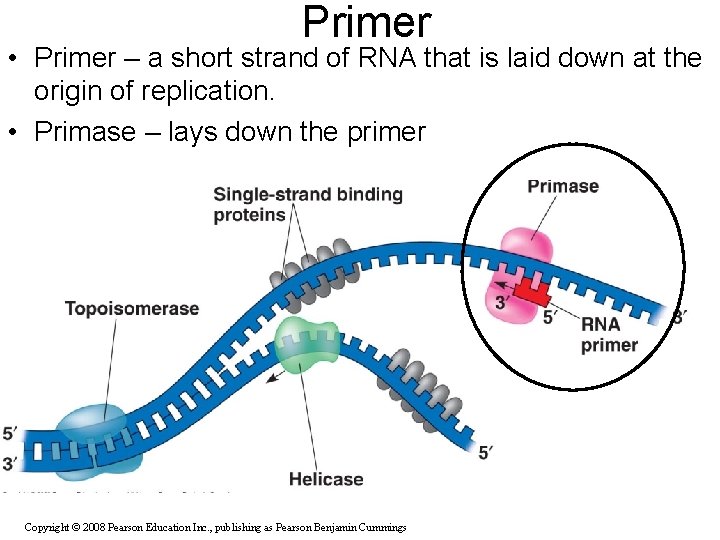
Primer • Primer – a short strand of RNA that is laid down at the origin of replication. • Primase – lays down the primer Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
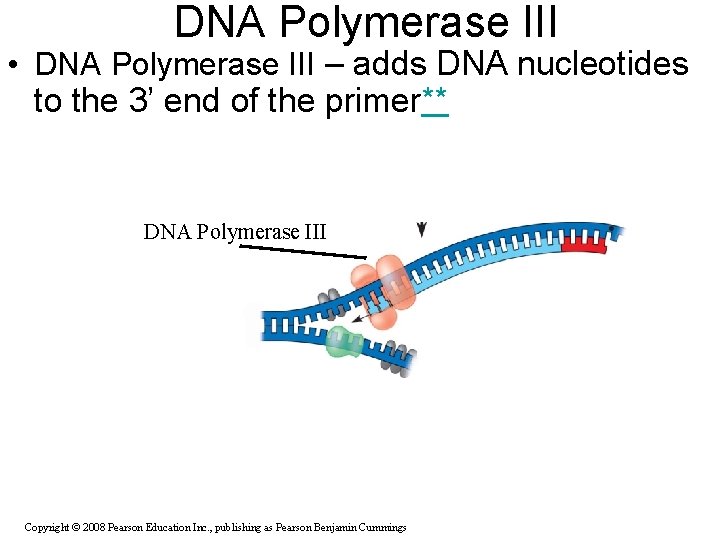
DNA Polymerase III • DNA Polymerase III – adds DNA nucleotides to the 3’ end of the primer** DNA Polymerase III Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
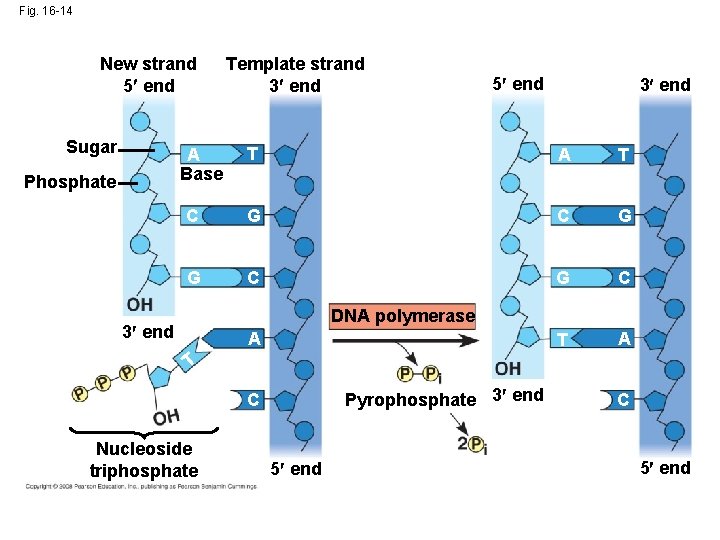
Fig. 16 -14 New strand 5 end Sugar 5 end 3 end T A T C G G C T A A Base Phosphate Template strand 3 end DNA polymerase 3 end A T Pyrophosphate 3 end C Nucleoside triphosphate 5 end C 5 end
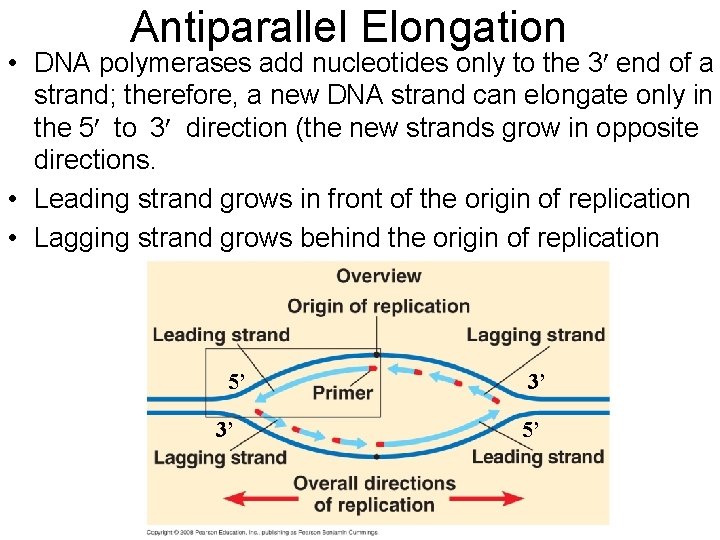
Antiparallel Elongation • DNA polymerases add nucleotides only to the 3 end of a strand; therefore, a new DNA strand can elongate only in the 5 to 3 direction (the new strands grow in opposite directions. • Leading strand grows in front of the origin of replication • Lagging strand grows behind the origin of replication 5’ 3’ 3’ 5’
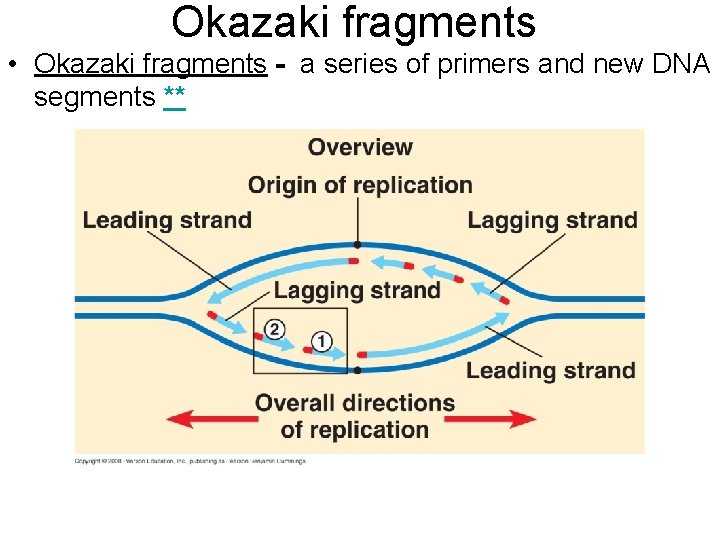
Okazaki fragments • Okazaki fragments - a series of primers and new DNA segments **
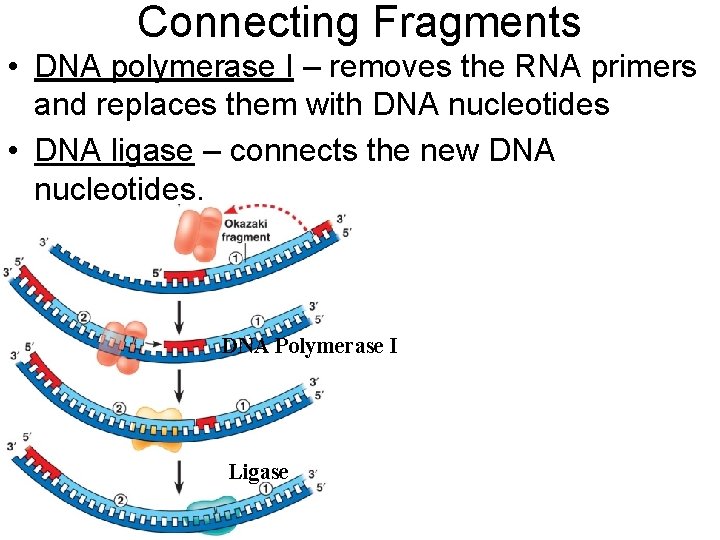
Connecting Fragments • DNA polymerase I – removes the RNA primers and replaces them with DNA nucleotides • DNA ligase – connects the new DNA nucleotides. DNA Polymerase I Ligase
Fig. 16 -16 b 1 3 Template strand 5 5 3
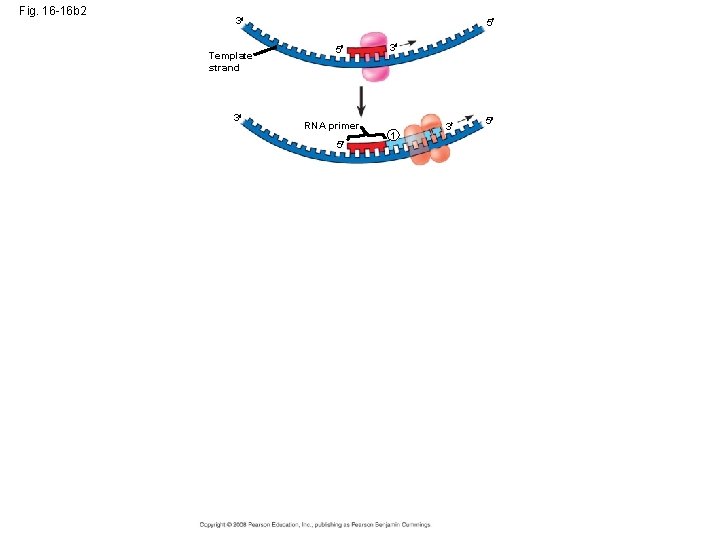
Fig. 16 -16 b 2 3 Template strand 3 5 5 RNA primer 5 3 1 3 5
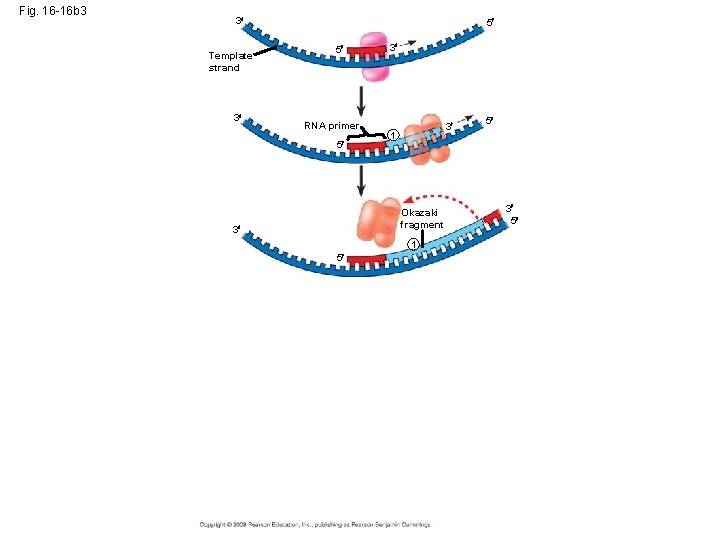
Fig. 16 -16 b 3 3 Template strand 3 5 5 RNA primer 5 3 3 1 Okazaki fragment 3 1 5 5 3 5
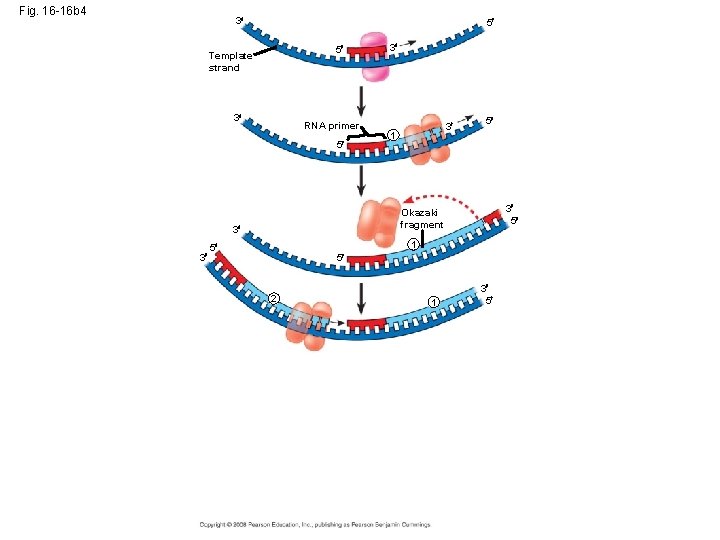
Fig. 16 -16 b 4 3 5 5 Template strand 3 RNA primer 5 3 1 5 3 5 Okazaki fragment 3 3 3 1 5 5 2 1 3 5
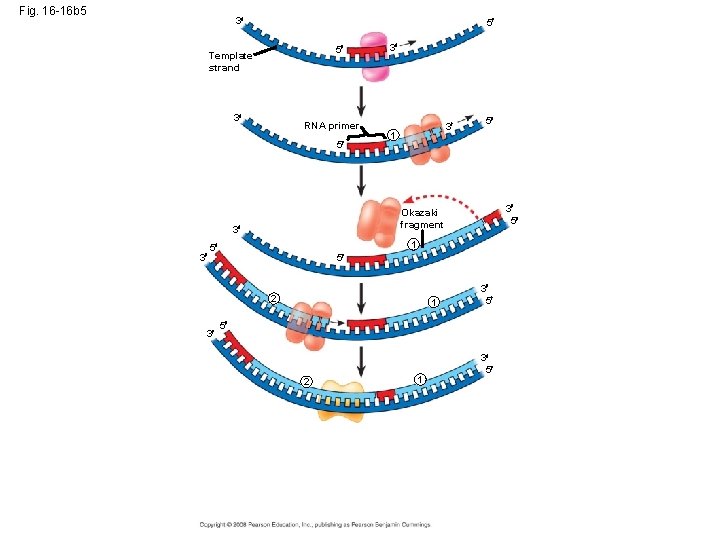
Fig. 16 -16 b 5 3 5 5 Template strand 3 RNA primer 5 3 1 3 5 1 5 5 2 3 5 Okazaki fragment 3 3 3 1 3 5 5 2 1 3 5
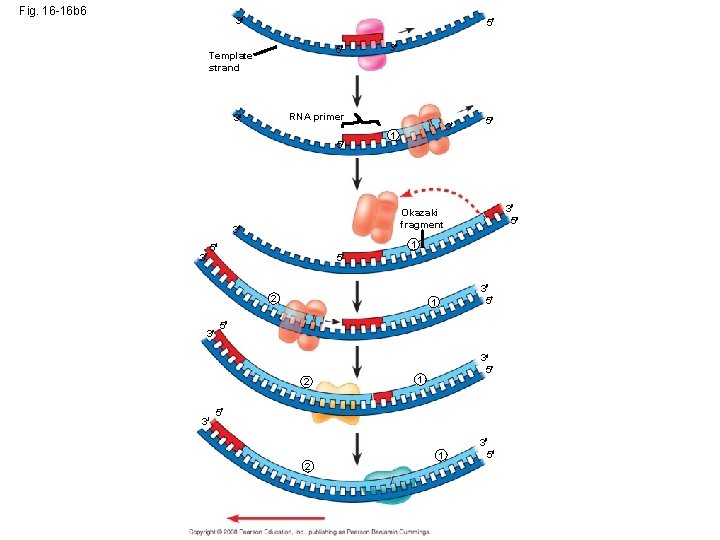
Fig. 16 -16 b 6 3 5 5 Template strand RNA primer 3 5 3 1 5 2 3 5 1 5 2 3 3 5 1 5 3 5 Okazaki fragment 3 3 3 3 5 1 5 2 1 3 5
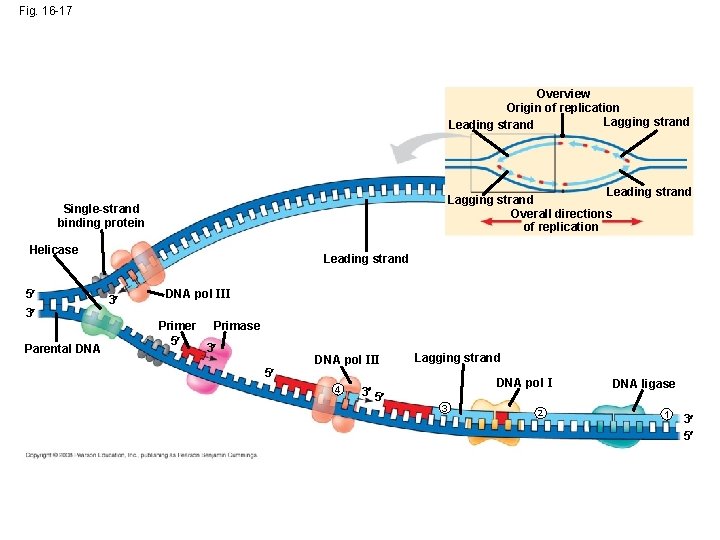
Fig. 16 -17 Overview Origin of replication Lagging strand Leading strand Lagging strand Overall directions of replication Single-strand binding protein Helicase 5 3 Parental DNA Leading strand 3 DNA pol III Primer 5 Primase 3 5 DNA pol III 4 3 5 Lagging strand DNA pol I 3 2 DNA ligase 1 3 5
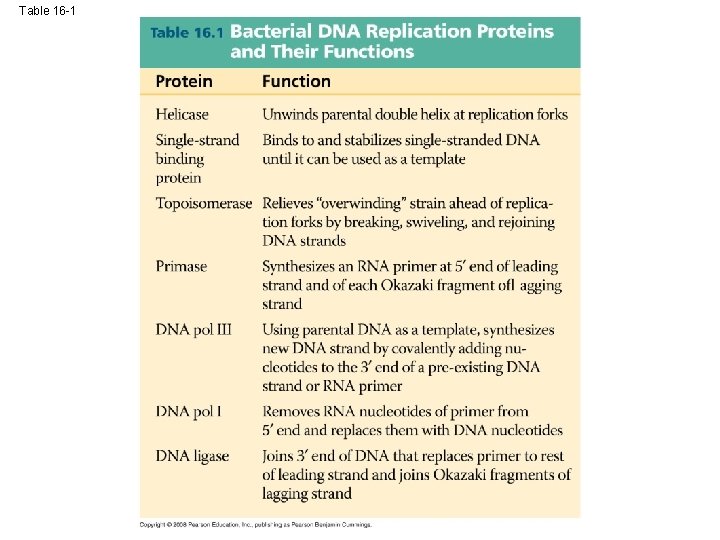
Table 16 -1
Proofreading and Repairing DNA • DNA Polymerases proofread DNA as it is added. • Enzymes can repair mismatched based pairs (mismatch repair) or damaged DNA (X-rays, UV, chemicals) Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
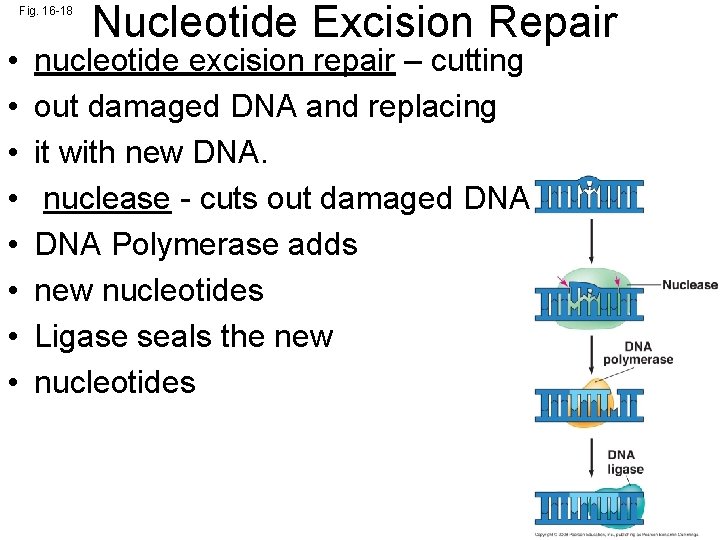
Fig. 16 -18 • • Nucleotide Excision Repair nucleotide excision repair – cutting out damaged DNA and replacing it with new DNA. nuclease - cuts out damaged DNA Polymerase adds new nucleotides Ligase seals the new nucleotides
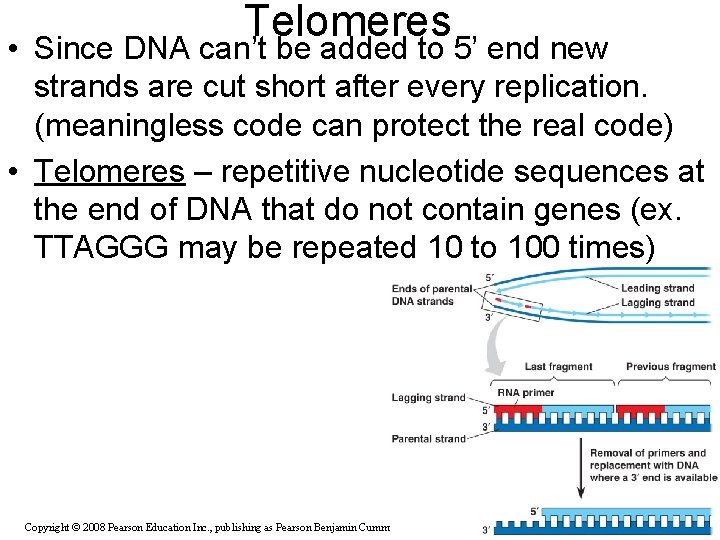
Telomeres • Since DNA can’t be added to 5’ end new strands are cut short after every replication. (meaningless code can protect the real code) • Telomeres – repetitive nucleotide sequences at the end of DNA that do not contain genes (ex. TTAGGG may be repeated 10 to 100 times) Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
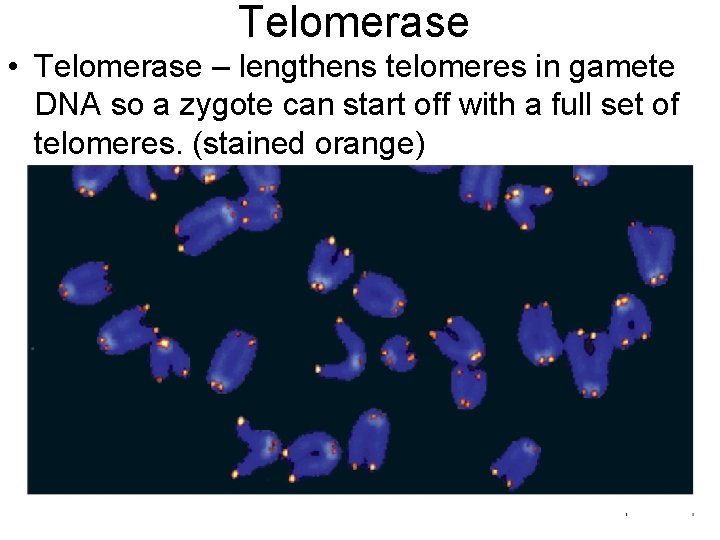
Telomerase • Telomerase – lengthens telomeres in gamete DNA so a zygote can start off with a full set of telomeres. (stained orange)
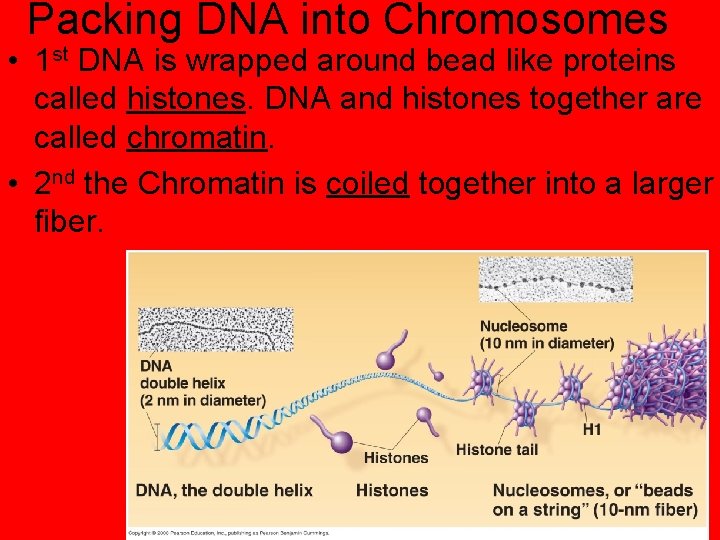
Packing DNA into Chromosomes • 1 st DNA is wrapped around bead like proteins called histones. DNA and histones together are called chromatin. • 2 nd the Chromatin is coiled together into a larger fiber.
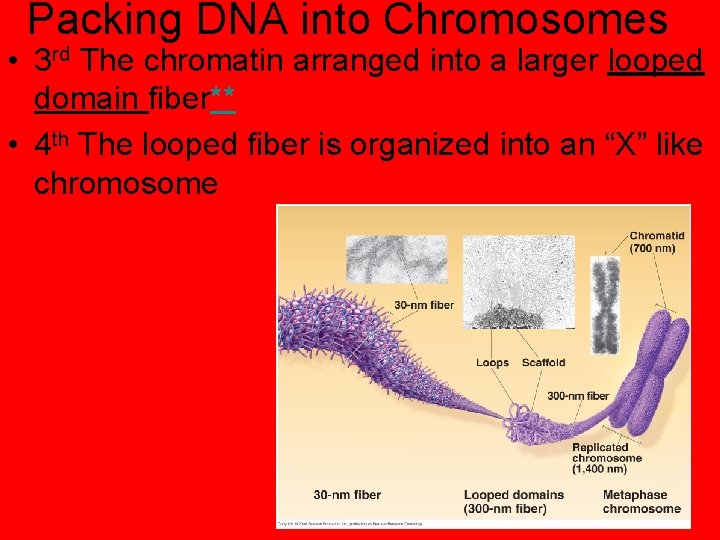
Packing DNA into Chromosomes • 3 rd The chromatin arranged into a larger looped domain fiber** • 4 th The looped fiber is organized into an “X” like chromosome
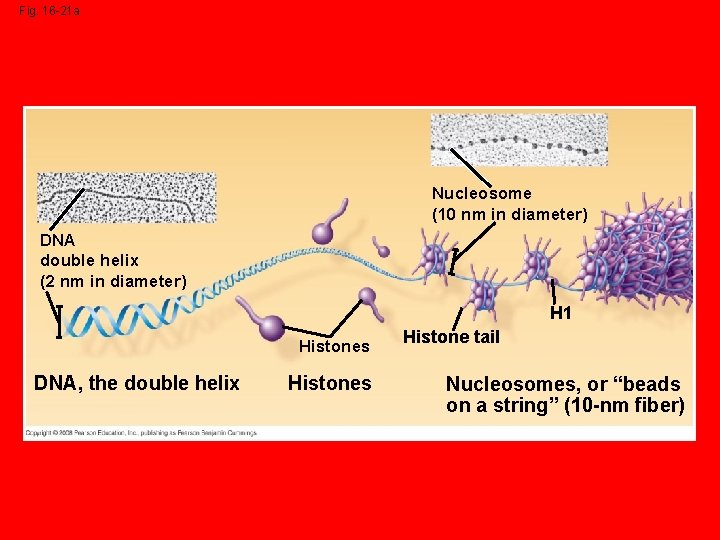
Fig. 16 -21 a Nucleosome (10 nm in diameter) DNA double helix (2 nm in diameter) H 1 Histones DNA, the double helix Histones Histone tail Nucleosomes, or “beads on a string” (10 -nm fiber)
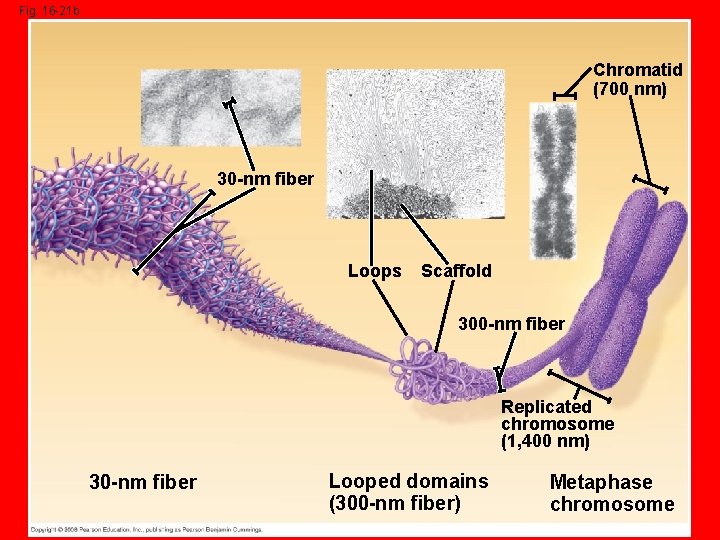
Fig. 16 -21 b Chromatid (700 nm) 30 -nm fiber Loops Scaffold 300 -nm fiber Replicated chromosome (1, 400 nm) 30 -nm fiber Looped domains (300 -nm fiber) Metaphase chromosome

You should now be able to: 1. Describe the structure of DNA 2. Describe the process of DNA replication; include the following terms: antiparallel structure, DNA polymerase, leading strand, lagging strand, Okazaki fragments, DNA ligase, primer, primase, helicase, topoisomerase, single-strand binding proteins 3. Describe DNA repair Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
4. Describe the function of telomeres 5. Compare a bacterial chromosome and a eukaryotic chromosome Copyright © 2008 Pearson Education Inc. , publishing as Pearson Benjamin Cummings
- Slides: 42