DNA Sequencing Steps to Assemble a Genome Some
DNA Sequencing
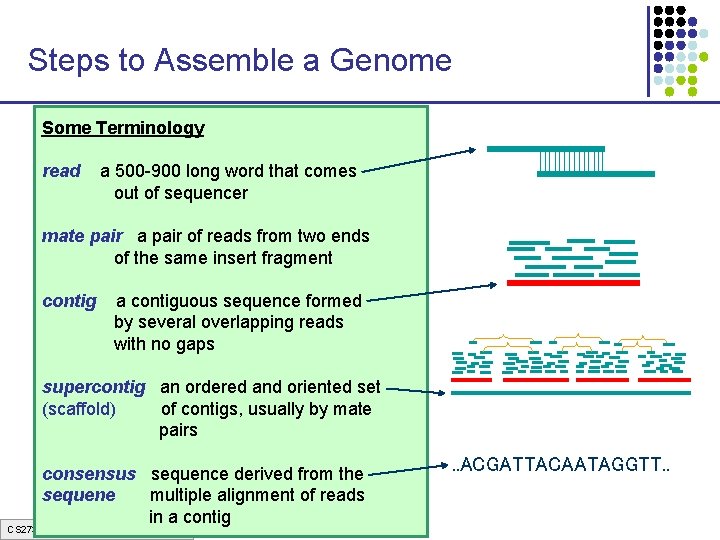
Steps to Assemble a Genome Some Terminology 1. Find overlapping readsthat comes read a 500 -900 long word out of sequencer mate pair a pair of reads from two ends 2. Merge some “good” of reads into of the same insert pairs fragment longer contigs contig a contiguous sequence formed by several overlapping reads with no gaps 3. Link contigs to form supercontigs supercontig an ordered and oriented set (scaffold) of contigs, usually by mate pairs 4. Derive consensus sequence derived from the sequene multiple alignment of reads in a contig CS 273 a Lecture 3, Spring 07, Batzoglou . . ACGATTACAATAGGTT. .

1. Find Overlapping Reads aaactgcagtacggatct aaactgcagt … gtacggatct gggcccaaactgcagtac gggcccaaac … actgcagtac gtacggatctactacaca gtacggatct … ctactacaca CS 273 a Lecture 3, Spring 07, Batzoglou (read, pos. , word, orient. ) (word, read, orient. , pos. ) aaactgcagt actgcagta … gtacggatct gggcccaaac gcccaaact … actgcagtac gtacggatct acggatcta … ctactacaca aaactgcagt acggatcta actgcagta cccaaactg cggatctactacac ctgcagtac gcccaaact ggcccaaac gggcccaaa gtacggatct tactacaca
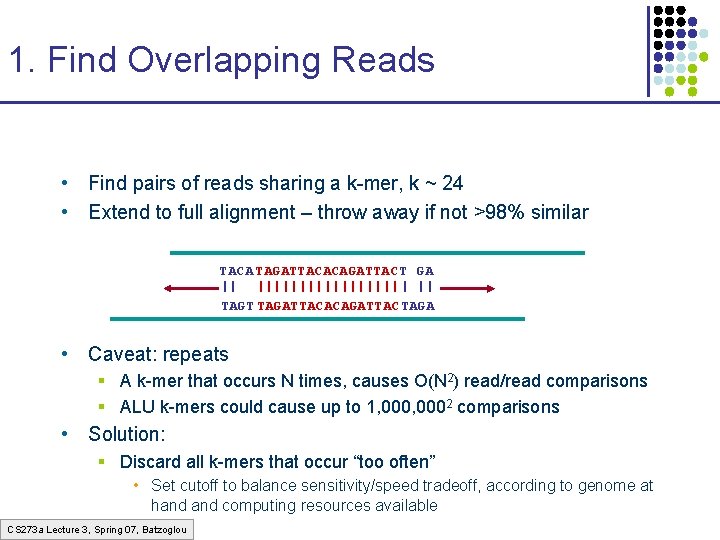
1. Find Overlapping Reads • Find pairs of reads sharing a k-mer, k ~ 24 • Extend to full alignment – throw away if not >98% similar TACA TAGATTACACAGATTAC T GA || ||||||||| | || TAGT TAGATTACACAGATTAC TAGA • Caveat: repeats § A k-mer that occurs N times, causes O(N 2) read/read comparisons § ALU k-mers could cause up to 1, 0002 comparisons • Solution: § Discard all k-mers that occur “too often” • Set cutoff to balance sensitivity/speed tradeoff, according to genome at hand computing resources available CS 273 a Lecture 3, Spring 07, Batzoglou

1. Find Overlapping Reads Create local multiple alignments from the overlapping reads TAGATTACACAGATTACTGA TAG TTACACAGATTATTGA TAGATTACACAGATTACTGA CS 273 a Lecture 3, Spring 07, Batzoglou
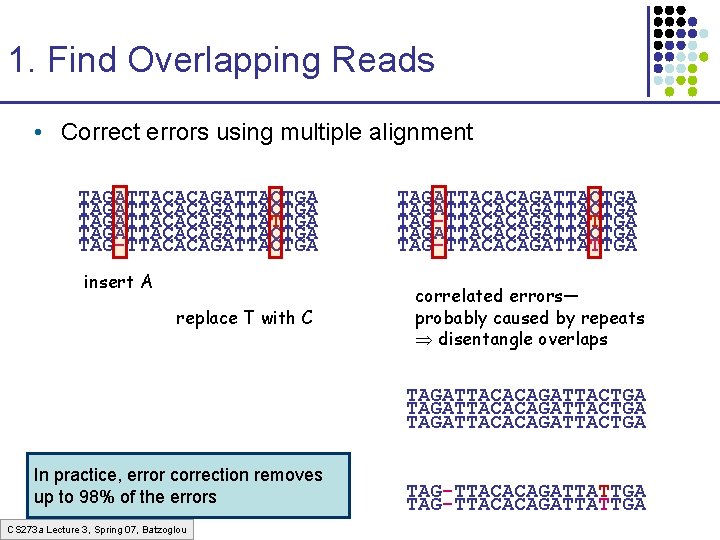
1. Find Overlapping Reads • Correct errors using multiple alignment TAGATTACACAGATTACTGA TAGATTACACAGATTATTGA TAGATTACACAGATTACTGA TAG-TTACACAGATTACTGA insert A replace T with C TAGATTACACAGATTACTGA TAG-TTACACAGATTATTGA correlated errors— probably caused by repeats disentangle overlaps TAGATTACACAGATTACTGA In practice, error correction removes up to 98% of the errors CS 273 a Lecture 3, Spring 07, Batzoglou TAG-TTACACAGATTATTGA
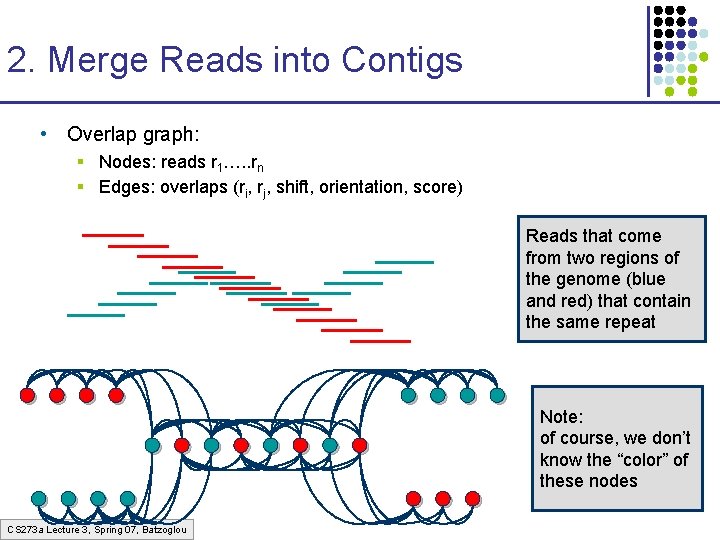
2. Merge Reads into Contigs • Overlap graph: § Nodes: reads r 1…. . rn § Edges: overlaps (ri, rj, shift, orientation, score) Reads that come from two regions of the genome (blue and red) that contain the same repeat Note: of course, we don’t know the “color” of these nodes CS 273 a Lecture 3, Spring 07, Batzoglou
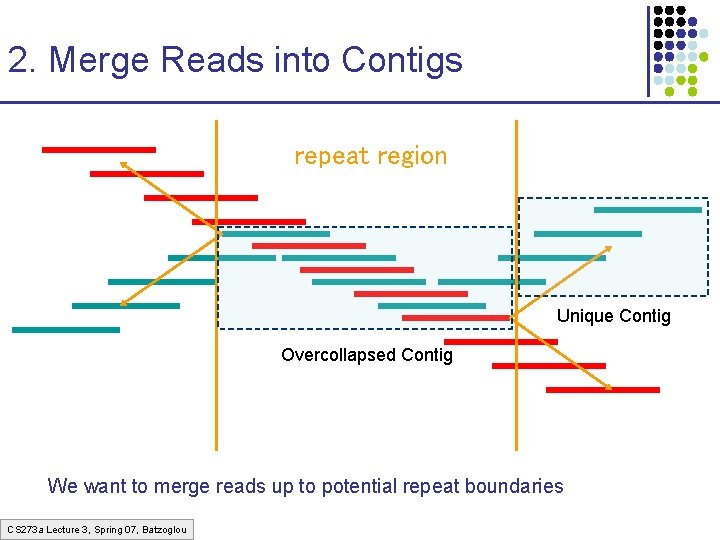
2. Merge Reads into Contigs repeat region Unique Contig Overcollapsed Contig We want to merge reads up to potential repeat boundaries CS 273 a Lecture 3, Spring 07, Batzoglou
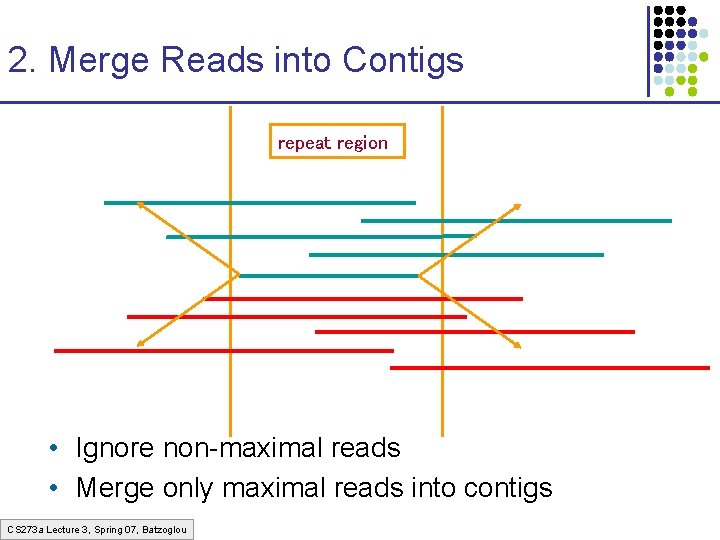
2. Merge Reads into Contigs repeat region • Ignore non-maximal reads • Merge only maximal reads into contigs CS 273 a Lecture 3, Spring 07, Batzoglou
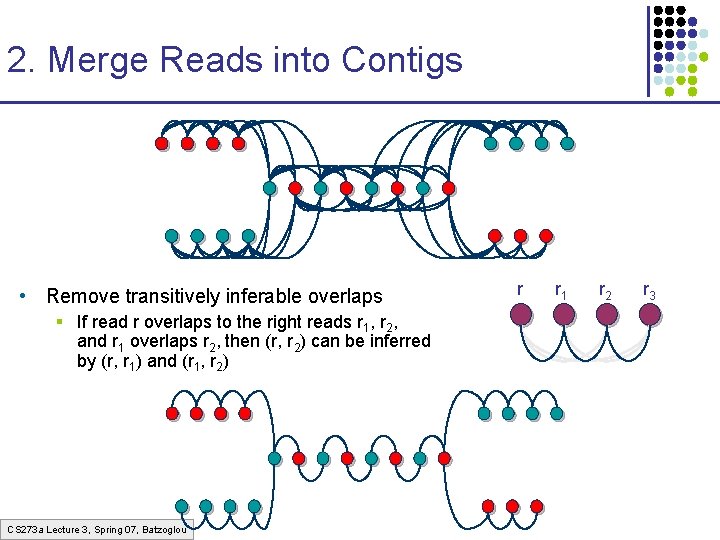
2. Merge Reads into Contigs • Remove transitively inferable overlaps § If read r overlaps to the right reads r 1, r 2, and r 1 overlaps r 2, then (r, r 2) can be inferred by (r, r 1) and (r 1, r 2) CS 273 a Lecture 3, Spring 07, Batzoglou r r 1 r 2 r 3
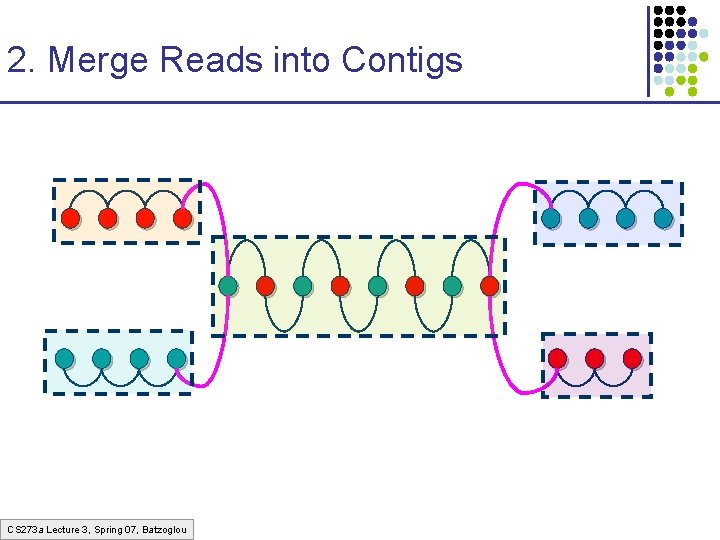
2. Merge Reads into Contigs CS 273 a Lecture 3, Spring 07, Batzoglou
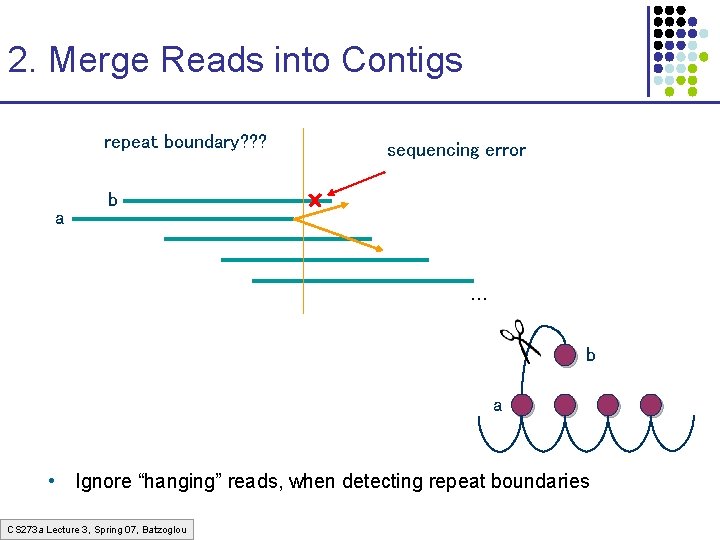
2. Merge Reads into Contigs repeat boundary? ? ? a sequencing error b … b a • Ignore “hanging” reads, when detecting repeat boundaries CS 273 a Lecture 3, Spring 07, Batzoglou
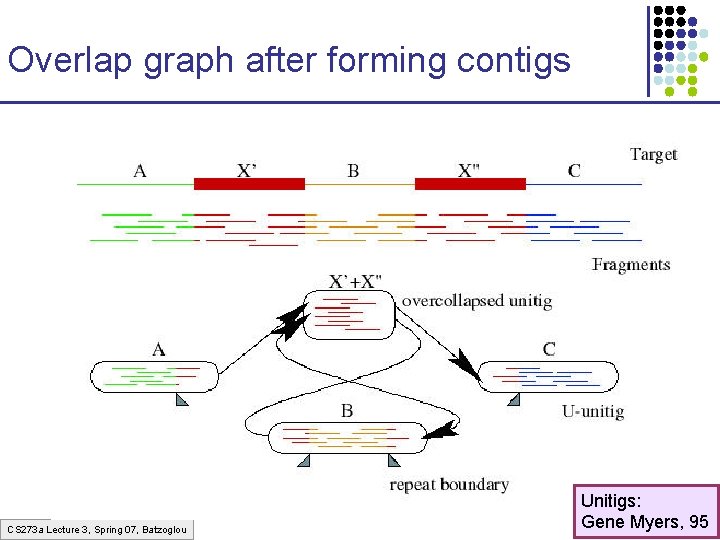
Overlap graph after forming contigs CS 273 a Lecture 3, Spring 07, Batzoglou Unitigs: Gene Myers, 95
Repeats, errors, and contig lengths • Repeats shorter than read length are easily resolved § Read that spans across a repeat disambiguates order of flanking regions • Repeats with more base pair diffs than sequencing error rate are OK § We throw overlaps between two reads in different copies of the repeat • To make the genome appear less repetitive, try to: § Increase read length § Decrease sequencing error rate Role of error correction: Discards up to 98% of single-letter sequencing errors decreases error rate decreases effective repeat content increases contig length CS 273 a Lecture 3, Spring 07, Batzoglou
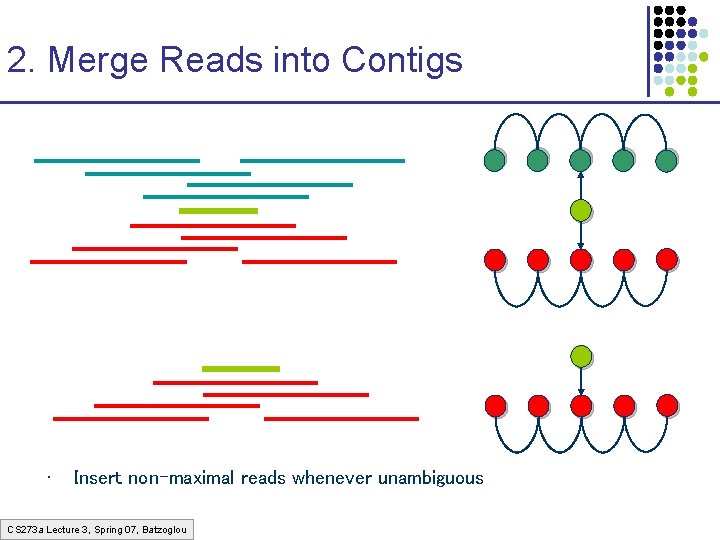
2. Merge Reads into Contigs • Insert non-maximal reads whenever unambiguous CS 273 a Lecture 3, Spring 07, Batzoglou
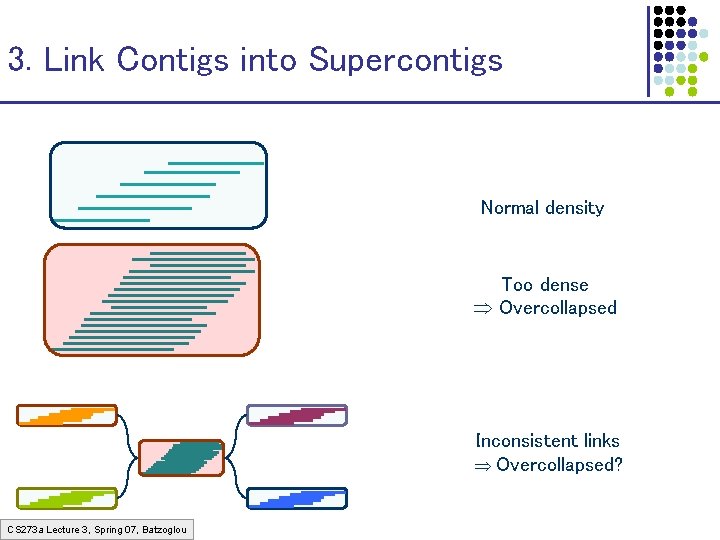
3. Link Contigs into Supercontigs Normal density Too dense Overcollapsed Inconsistent links Overcollapsed? CS 273 a Lecture 3, Spring 07, Batzoglou
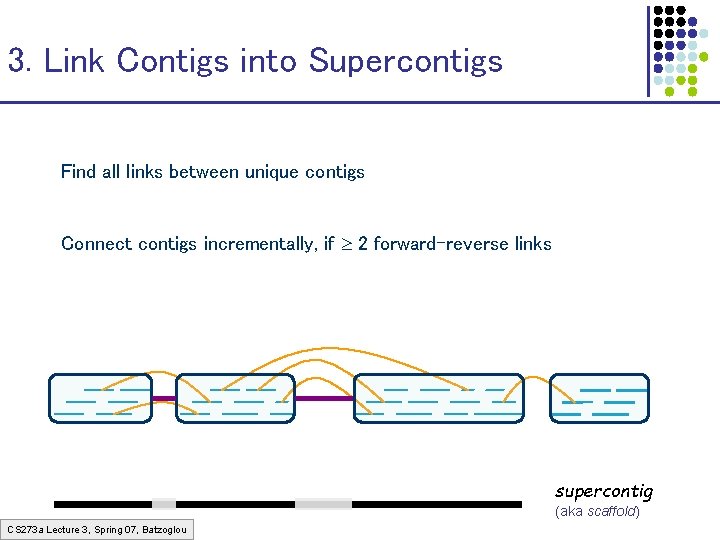
3. Link Contigs into Supercontigs Find all links between unique contigs Connect contigs incrementally, if 2 forward-reverse links supercontig (aka scaffold) CS 273 a Lecture 3, Spring 07, Batzoglou
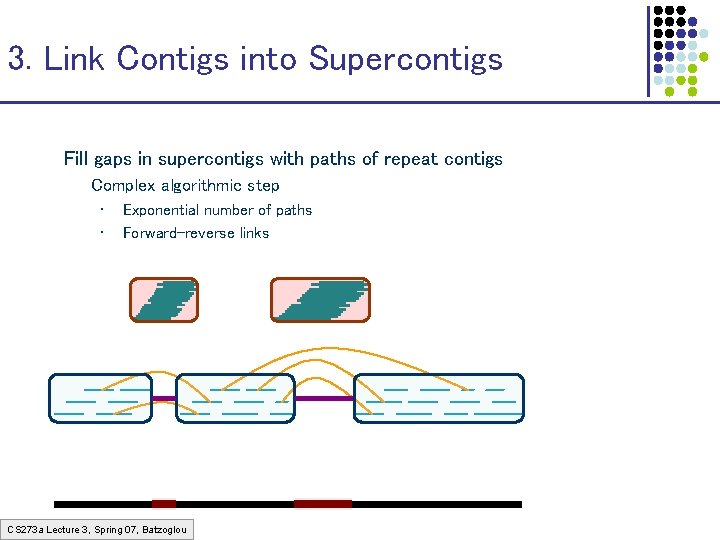
3. Link Contigs into Supercontigs Fill gaps in supercontigs with paths of repeat contigs Complex algorithmic step • • Exponential number of paths Forward-reverse links CS 273 a Lecture 3, Spring 07, Batzoglou
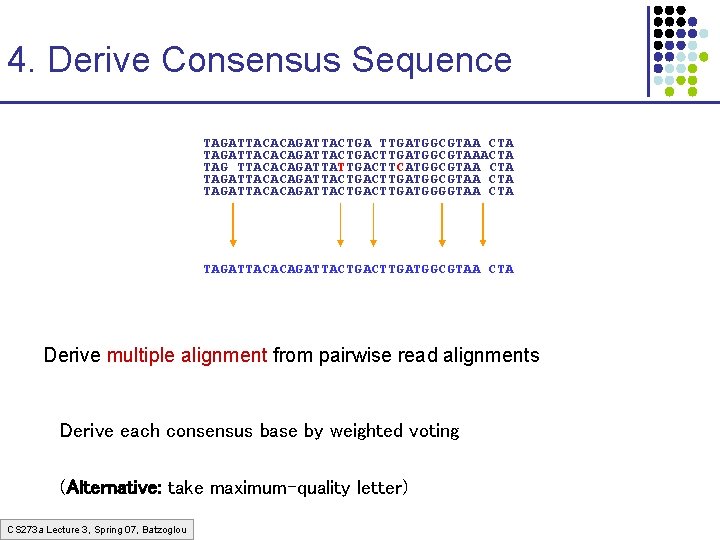
4. Derive Consensus Sequence TAGATTACACAGATTACTGA TTGATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAAACTA TAG TTACACAGATTATTGACTTCATGGCGTAA CTA TAGATTACACAGATTACTGACTTGATGGGGTAA CTA TAGATTACACAGATTACTGACTTGATGGCGTAA CTA Derive multiple alignment from pairwise read alignments Derive each consensus base by weighted voting (Alternative: take maximum-quality letter) CS 273 a Lecture 3, Spring 07, Batzoglou
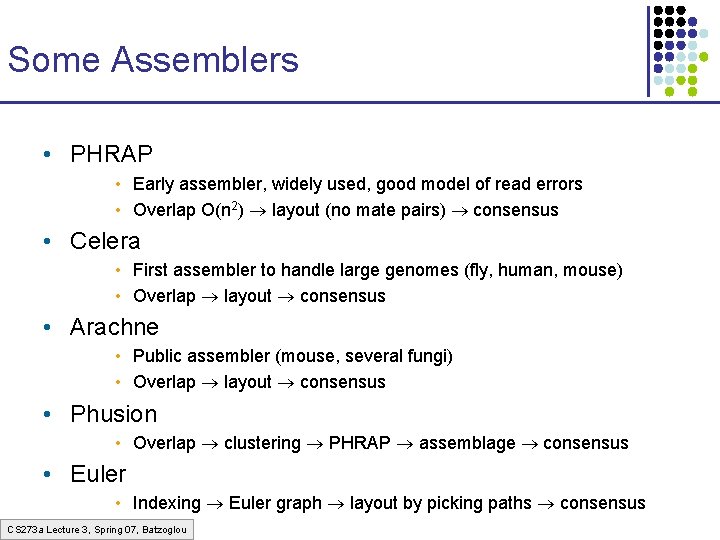
Some Assemblers • PHRAP • Early assembler, widely used, good model of read errors • Overlap O(n 2) layout (no mate pairs) consensus • Celera • First assembler to handle large genomes (fly, human, mouse) • Overlap layout consensus • Arachne • Public assembler (mouse, several fungi) • Overlap layout consensus • Phusion • Overlap clustering PHRAP assemblage consensus • Euler • Indexing Euler graph layout by picking paths consensus CS 273 a Lecture 3, Spring 07, Batzoglou
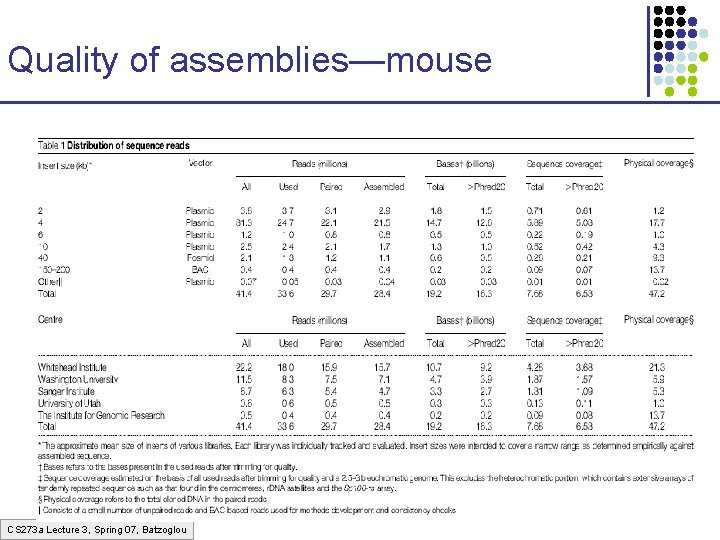
Quality of assemblies—mouse CS 273 a Lecture 3, Spring 07, Batzoglou
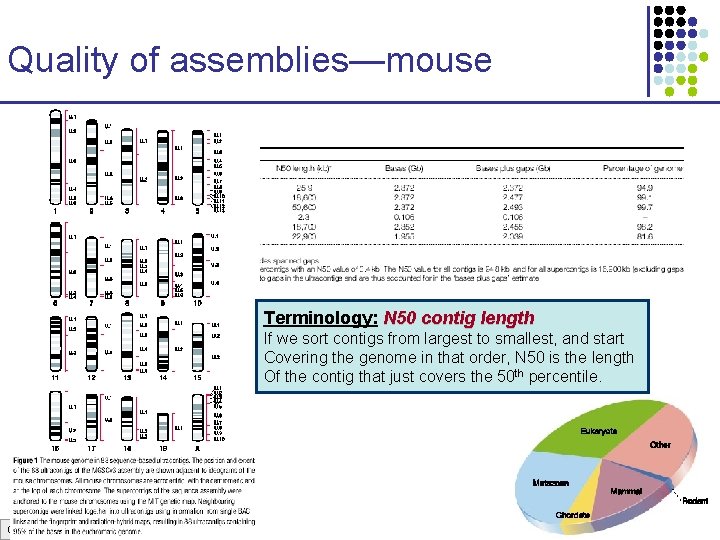
Quality of assemblies—mouse Terminology: N 50 contig length If we sort contigs from largest to smallest, and start Covering the genome in that order, N 50 is the length Of the contig that just covers the 50 th percentile. CS 273 a Lecture 3, Spring 07, Batzoglou
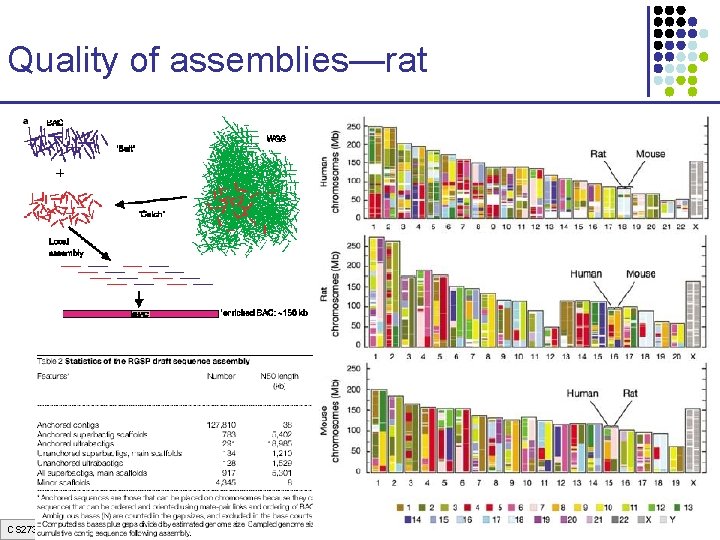
Quality of assemblies—rat CS 273 a Lecture 3, Spring 07, Batzoglou
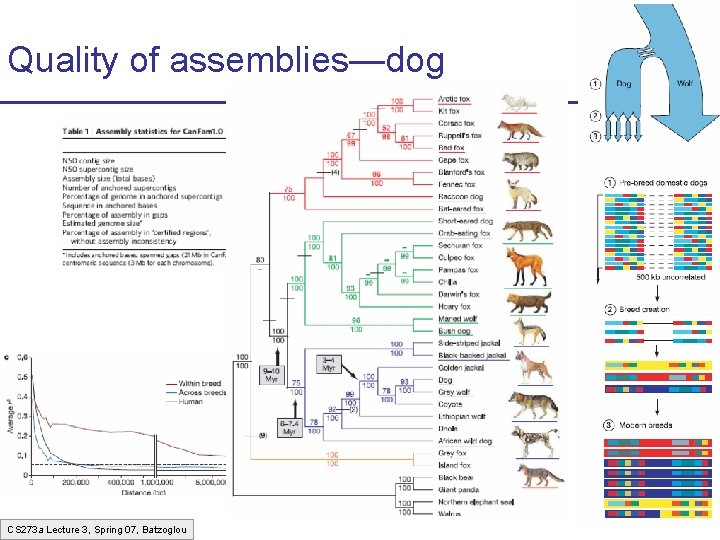
Quality of assemblies—dog CS 273 a Lecture 3, Spring 07, Batzoglou
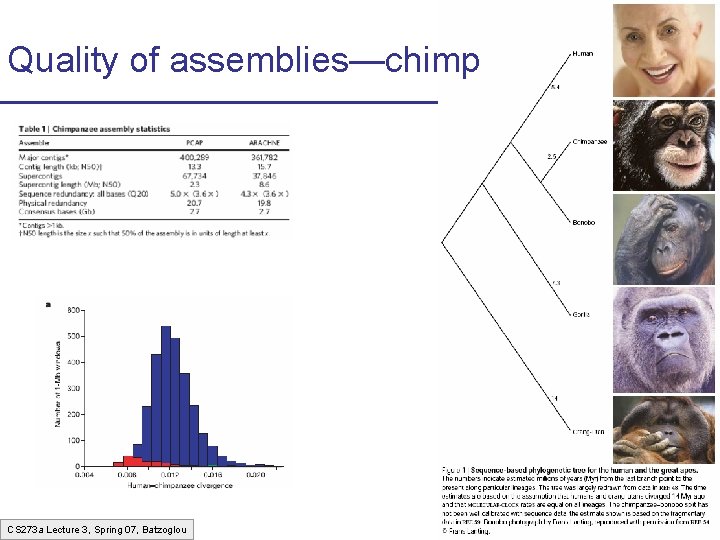
Quality of assemblies—chimp CS 273 a Lecture 3, Spring 07, Batzoglou

History of WGA 1997 • 1982: -virus, 48, 502 bp • 1995: h-influenzae, Let’s sequence the human 1 genome Mbp with the shotgun strategy • 2000: fly, 100 Mbp • 2001 – present Thatrat is*, chicken, dog, chimpanzee, § human (3 Gbp), mouse (2. 5 Gbp), several fungal genomes impossible, and a bad idea anyway Gene Myers CS 273 a Lecture 3, Spring 07, Batzoglou Phil Green

Some new sequencing technologies CS 273 a Lecture 3, Spring 07, Batzoglou
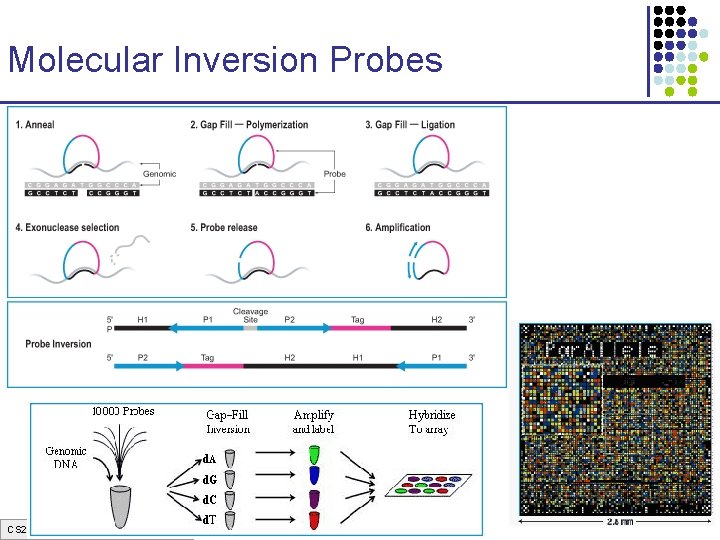
Molecular Inversion Probes CS 273 a Lecture 3, Spring 07, Batzoglou

Single Molecule Array for Genotyping—Solexa CS 273 a Lecture 3, Spring 07, Batzoglou
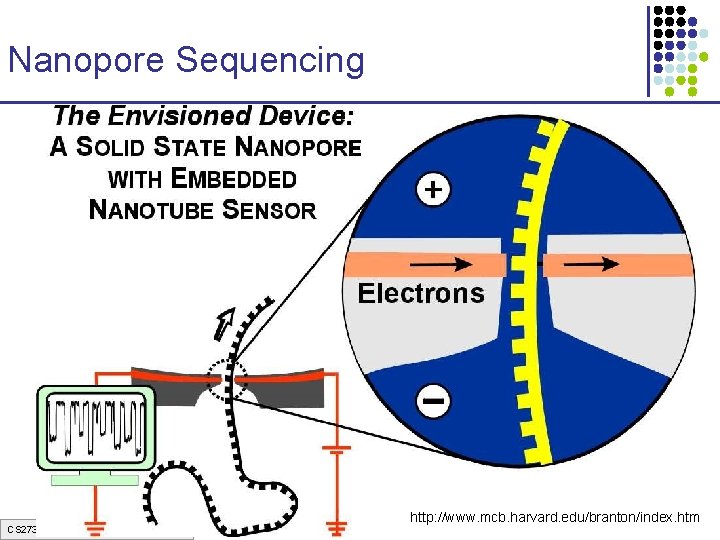
Nanopore Sequencing CS 273 a Lecture 3, Spring 07, Batzoglou http: //www. mcb. harvard. edu/branton/index. htm
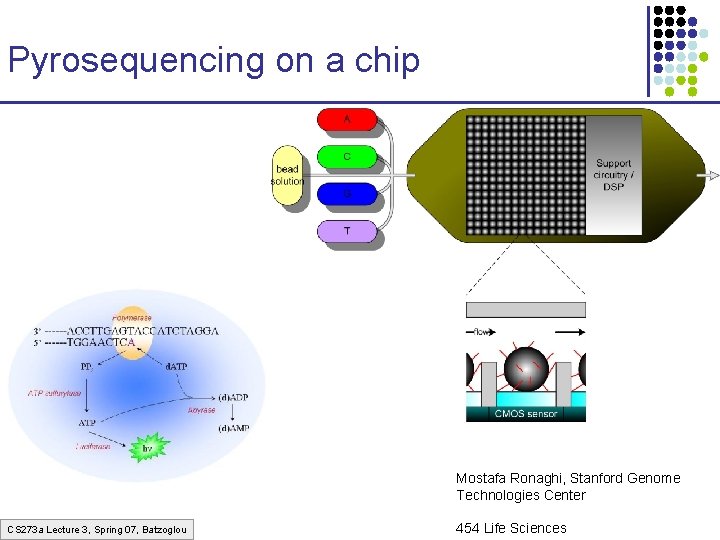
Pyrosequencing on a chip Mostafa Ronaghi, Stanford Genome Technologies Center CS 273 a Lecture 3, Spring 07, Batzoglou 454 Life Sciences
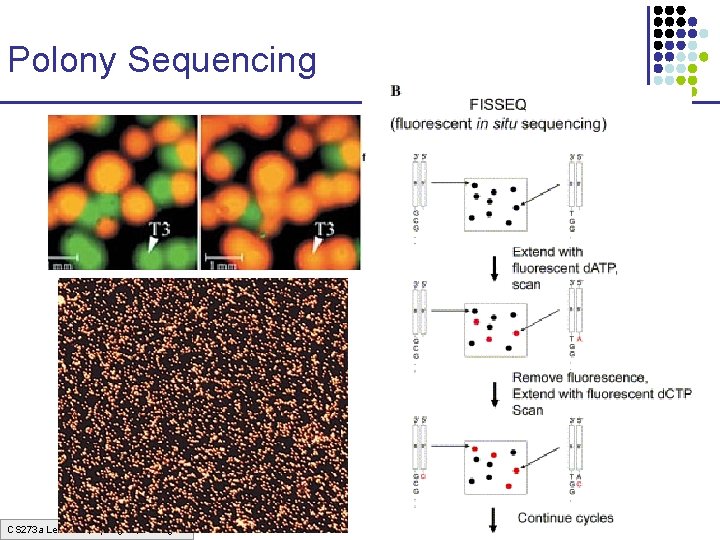
Polony Sequencing CS 273 a Lecture 3, Spring 07, Batzoglou
Some future directions for sequencing 1. Personalized genome sequencing • • Find your ~3, 000 single nucleotide polymorphisms (SNPs) Find your rearrangements • Goals: • • Link genome with phenotype Provide personalized diet and medicine (? ? ? ) designer babies, big-brother insurance companies Timeline: • • • Inexpensive sequencing: Genotype–phenotype association: Personalized drugs: CS 273 a Lecture 3, Spring 07, Batzoglou 2010 -2015 2010 -? ? ? 2015 -? ? ?
Some future directions for sequencing 2. Environmental sequencing • Find your flora: • • • External organs: skin, mucous membranes Gut, mouth, etc. Normal flora: >200 species, >trillions of individuals Flora–disease, flora–non-optimal health associations Timeline: • • organisms living in your body Inexpensive research sequencing: Research & associations Personalized sequencing today within next 10 years 2015+ Find diversity of organisms living in different environments • • Hard to isolate Assembly of all organisms at once CS 273 a Lecture 3, Spring 07, Batzoglou
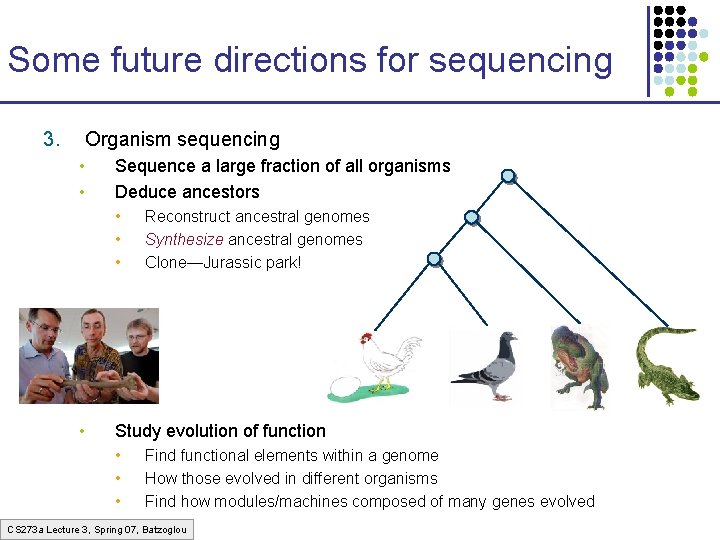
Some future directions for sequencing 3. Organism sequencing • • Sequence a large fraction of all organisms Deduce ancestors • • Reconstruct ancestral genomes Synthesize ancestral genomes Clone—Jurassic park! Study evolution of function • • • Find functional elements within a genome How those evolved in different organisms Find how modules/machines composed of many genes evolved CS 273 a Lecture 3, Spring 07, Batzoglou
- Slides: 35