DNA Sequencing Scenario DNA replication and chain termination
- Slides: 7

DNA Sequencing Scenario

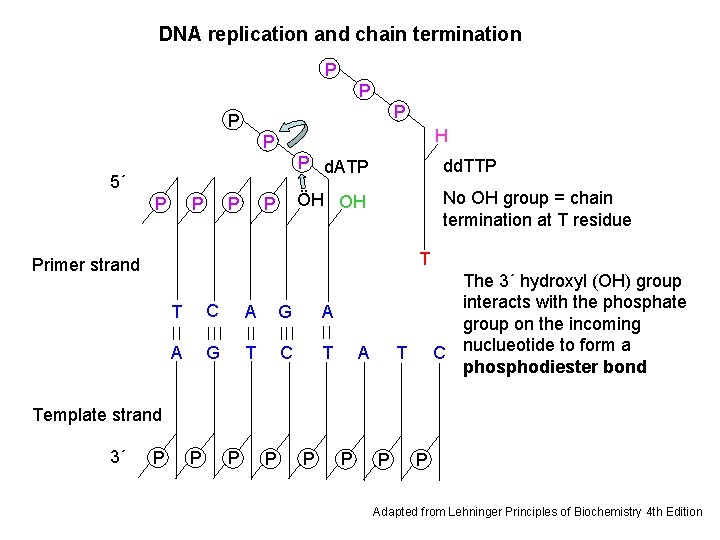
DNA replication and chain termination P P H P 5´ P P P d. ATP dd. TTP ÖH OH No OH group = chain termination at T residue T Primer strand T C A G A A G T C T A The 3´ hydroxyl (OH) group interacts with the phosphate group on the incoming C nuclueotide to form a phosphodiester bond T Template strand 3´ P P P P Adapted from Lehninger Principles of Biochemistry 4 th Edition

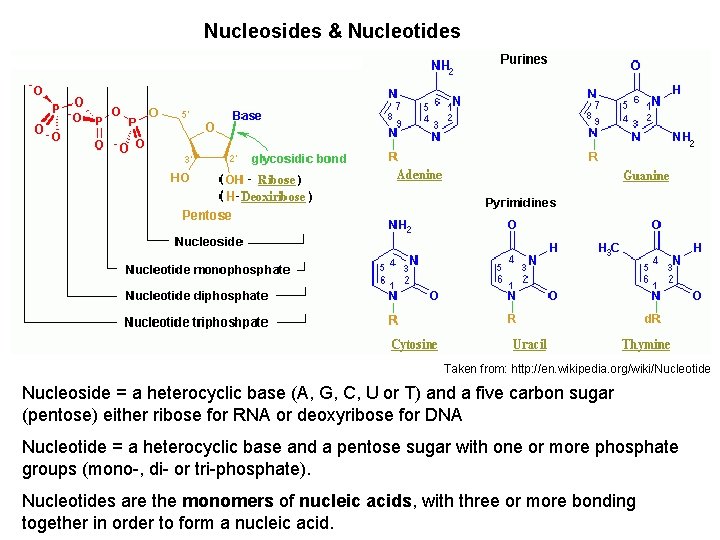
Nucleosides & Nucleotides Taken from: http: //en. wikipedia. org/wiki/Nucleotide Nucleoside = a heterocyclic base (A, G, C, U or T) and a five carbon sugar (pentose) either ribose for RNA or deoxyribose for DNA Nucleotide = a heterocyclic base and a pentose sugar with one or more phosphate groups (mono-, di- or tri-phosphate). Nucleotides are the monomers of nucleic acids, with three or more bonding together in order to form a nucleic acid.

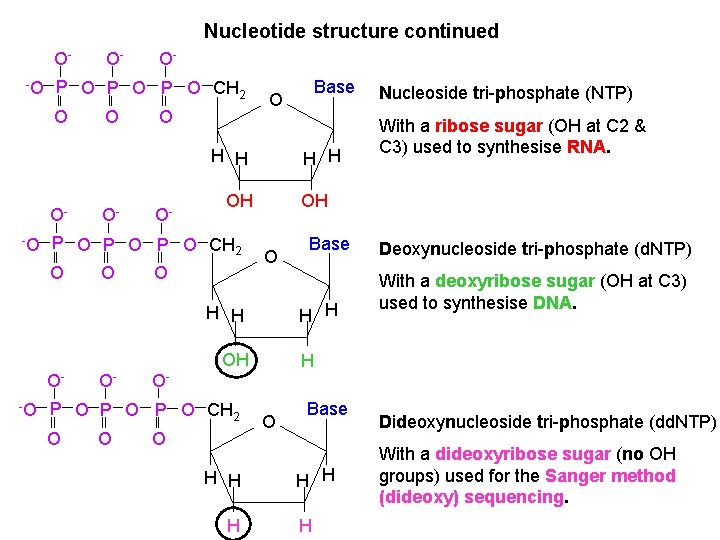
Nucleotide structure continued O-O O- O- P O P O CH 2 O O O H H O-O O- O- O O H H OH P O P O CH 2 O O -O O With a ribose sugar (OH at C 2 & C 3) used to synthesise RNA. Deoxynucleoside tri-phosphate (d. NTP) With a deoxyribose sugar (OH at C 3) used to synthesise DNA. H O- P O P O CH 2 O Base H H OH O- Nucleoside tri-phosphate (NTP) OH H H O- Base O O H H H O Base H H H Dideoxynucleoside tri-phosphate (dd. NTP) With a dideoxyribose sugar (no OH groups) used for the Sanger method (dideoxy) sequencing.

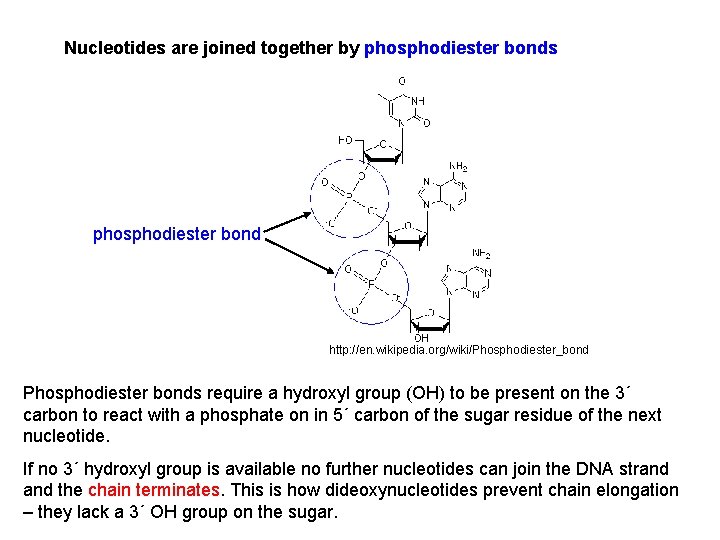
Nucleotides are joined together by phosphodiester bonds phosphodiester bond http: //en. wikipedia. org/wiki/Phosphodiester_bond Phosphodiester bonds require a hydroxyl group (OH) to be present on the 3´ carbon to react with a phosphate on in 5´ carbon of the sugar residue of the next nucleotide. If no 3´ hydroxyl group is available no further nucleotides can join the DNA strand the chain terminates. This is how dideoxynucleotides prevent chain elongation – they lack a 3´ OH group on the sugar.

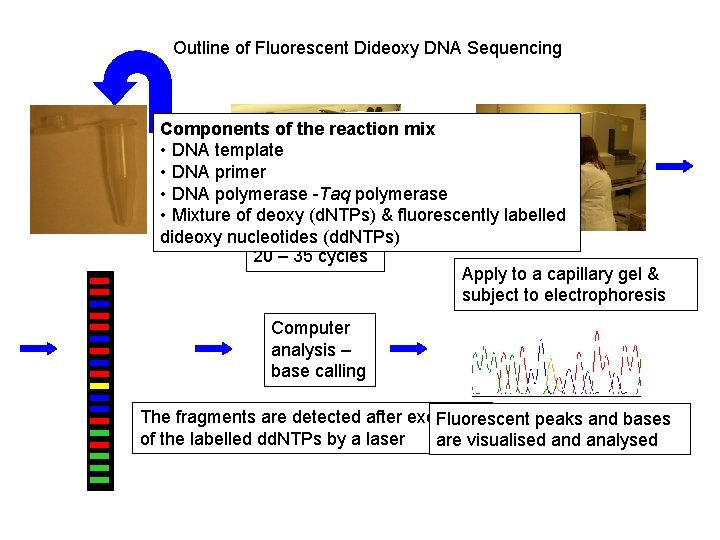
Outline of Fluorescent Dideoxy DNA Sequencing Components of the reaction mix • DNA template • DNA primer • DNA polymerase -Taq polymerase • Mixture of deoxy (d. NTPs) & fluorescently labelled dideoxy nucleotides (dd. NTPs) 20 – 35 cycles Apply to a capillary gel & subject to electrophoresis Computer analysis – base calling The fragments are detected after excitation Fluorescent peaks and bases of the labelled dd. NTPs by a laser are visualised analysed

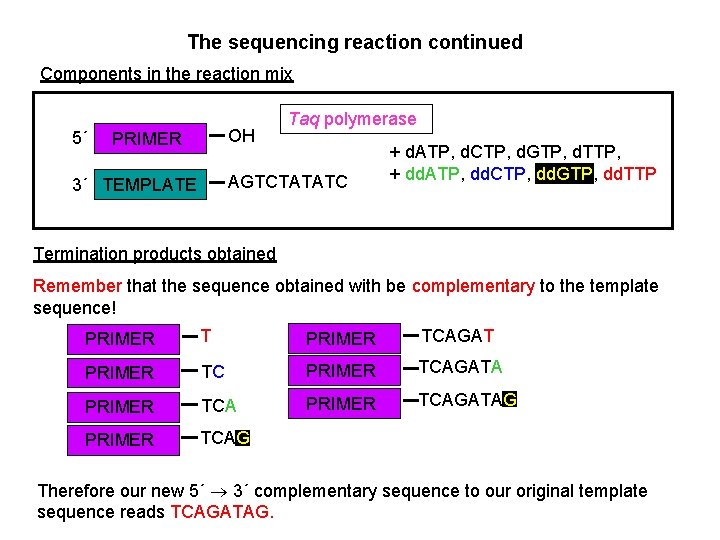
The sequencing reaction continued Components in the reaction mix 5´ OH PRIMER Taq polymerase AGTCTATATC 3´ TEMPLATE + d. ATP, d. CTP, d. GTP, d. TTP, + dd. ATP, dd. CTP, dd. GTP, dd. TTP Termination products obtained Remember that the sequence obtained with be complementary to the template sequence! PRIMER TCAGATA PRIMER TCAGATAG PRIMER TCAG Therefore our new 5´ 3´ complementary sequence to our original template sequence reads TCAGATAG.