DNA ReplicationII 1 03012022 Overview I General Features


- Slides: 15

DNA Replication-II 1 03/01/2022

Overview I. General Features of Replication • • Semi-Conservative Starts at Origin Bidirectional Semi-Discontinuous II. Identifying Proteins and Enzymes of Replication III. Detailed Examination of the Mechanism of Replication • Initiation • Priming • Elongation • Proofreading and Termination 2 03/01/2022

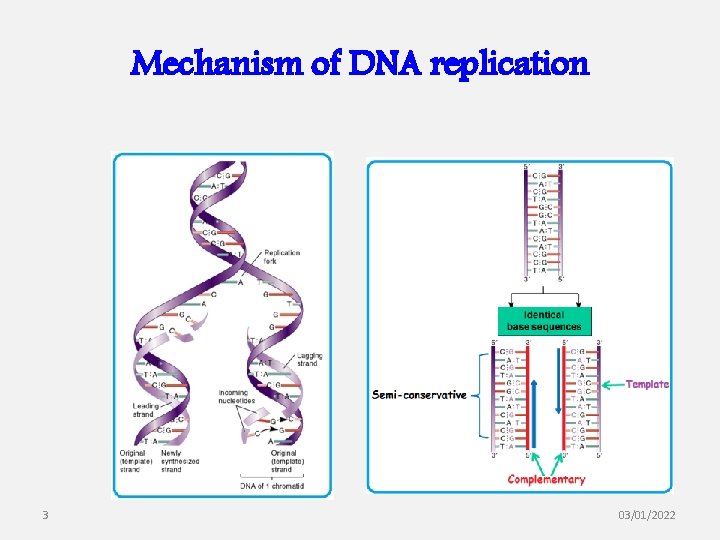
Mechanism of DNA replication 3 03/01/2022

DNA replication system • Template: Double stranded DNA • Substrate: d. NTP • Primer: Short RNA fragment with a free 3´-OH end • Enzyme: DNA-dependent DNA polymerase (DDDP), other enzymes, protein factor. 4 03/01/2022

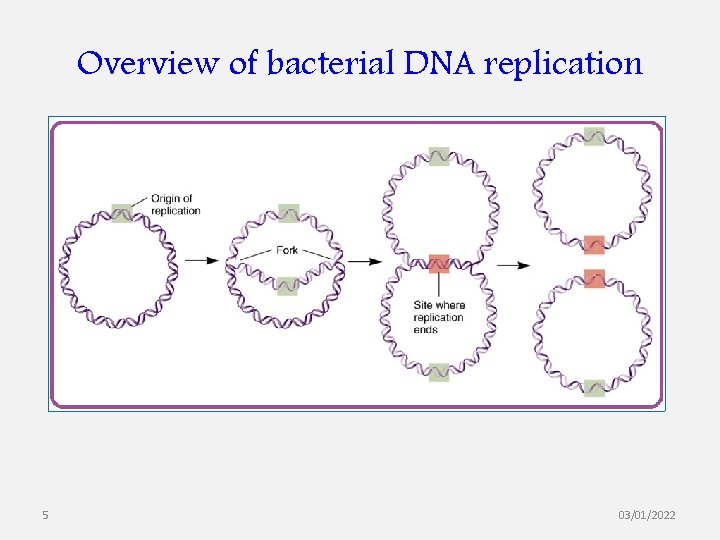
Overview of bacterial DNA replication 5 03/01/2022

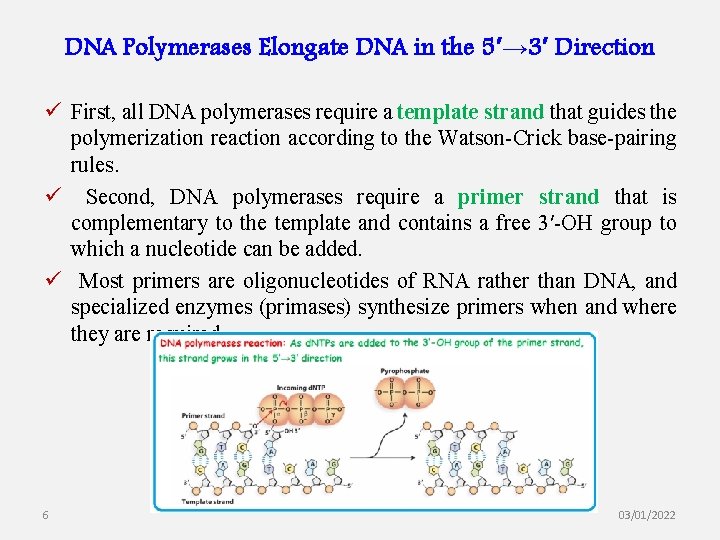
DNA Polymerases Elongate DNA in the 5′→ 3′ Direction ü First, all DNA polymerases require a template strand that guides the polymerization reaction according to the Watson-Crick base-pairing rules. ü Second, DNA polymerases require a primer strand that is complementary to the template and contains a free 3′-OH group to which a nucleotide can be added. ü Most primers are oligonucleotides of RNA rather than DNA, and specialized enzymes (primases) synthesize primers when and where they are required. 6 03/01/2022

DNA polymerases can only add nucleotides to a preexisting strand; they cannot synthesize DNA starting from only a template strand. Many different DNA polymerases have been studied from various types of bacteria, archaea, and eukaryotes, and mitochondria from many different species. All known DNA polymerases use the mechanism. 7 03/01/2022

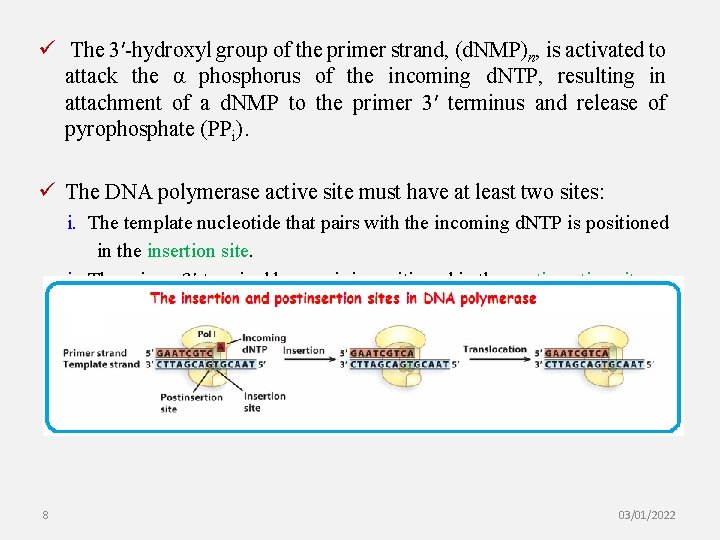
ü The 3′-hydroxyl group of the primer strand, (d. NMP)n, is activated to attack the α phosphorus of the incoming d. NTP, resulting in attachment of a d. NMP to the primer 3′ terminus and release of pyrophosphate (PPi). ü The DNA polymerase active site must have at least two sites: i. The template nucleotide that pairs with the incoming d. NTP is positioned in the insertion site. i. The primer 3′-terminal base pair is positioned in the postinsertion site. 8 03/01/2022

9

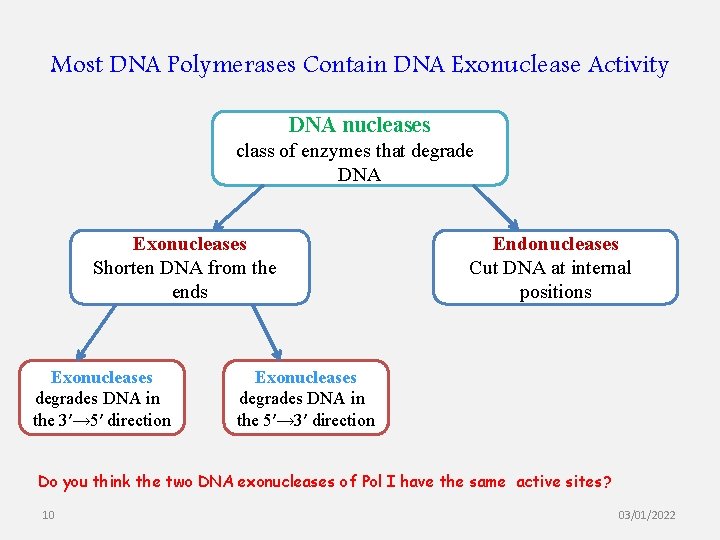
Most DNA Polymerases Contain DNA Exonuclease Activity DNA nucleases class of enzymes that degrade DNA Exonucleases Shorten DNA from the ends Exonucleases degrades DNA in the 3′→ 5′ direction Endonucleases Cut DNA at internal positions Exonucleases degrades DNA in the 5′→ 3′ direction Do you think the two DNA exonucleases of Pol I have the same active sites? 10 03/01/2022

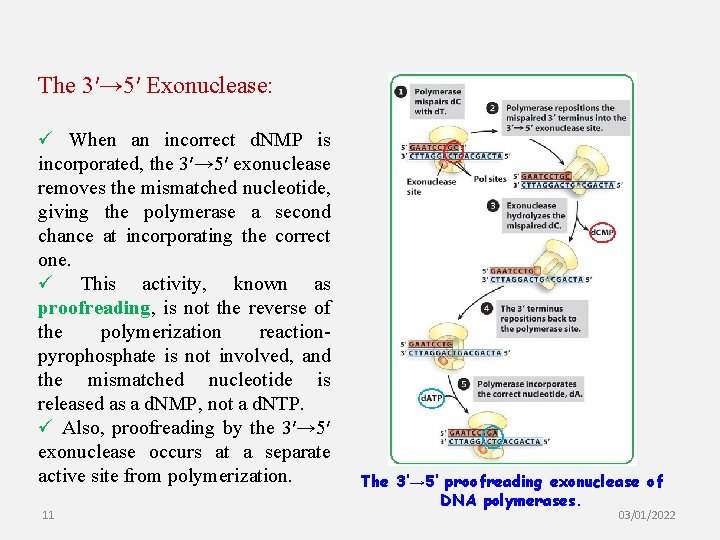
The 3′→ 5′ Exonuclease: ü When an incorrect d. NMP is incorporated, the 3′→ 5′ exonuclease removes the mismatched nucleotide, giving the polymerase a second chance at incorporating the correct one. ü This activity, known as proofreading, is not the reverse of the polymerization reactionpyrophosphate is not involved, and the mismatched nucleotide is released as a d. NMP, not a d. NTP. ü Also, proofreading by the 3′→ 5′ exonuclease occurs at a separate active site from polymerization. 11 The 3′→ 5′ proofreading exonuclease of DNA polymerases. 03/01/2022

Most DNA polymerases contain this proofreading exonuclease activity, although some do not and thus become more capable of bypassing a DNA lesion. 12 03/01/2022

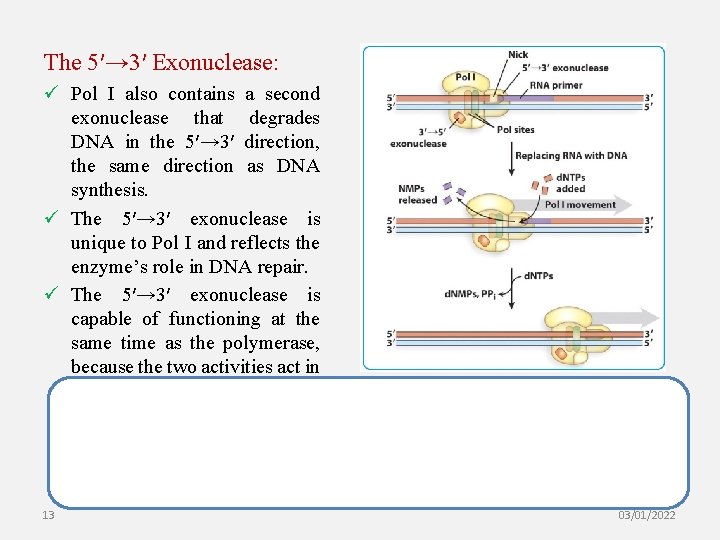
The 5′→ 3′ Exonuclease: ü Pol I also contains a second exonuclease that degrades DNA in the 5′→ 3′ direction, the same direction as DNA synthesis. ü The 5′→ 3′ exonuclease is unique to Pol I and reflects the enzyme’s role in DNA repair. ü The 5′→ 3′ exonuclease is capable of functioning at the same time as the polymerase, because the two activities act in the same direction. 13 03/01/2022

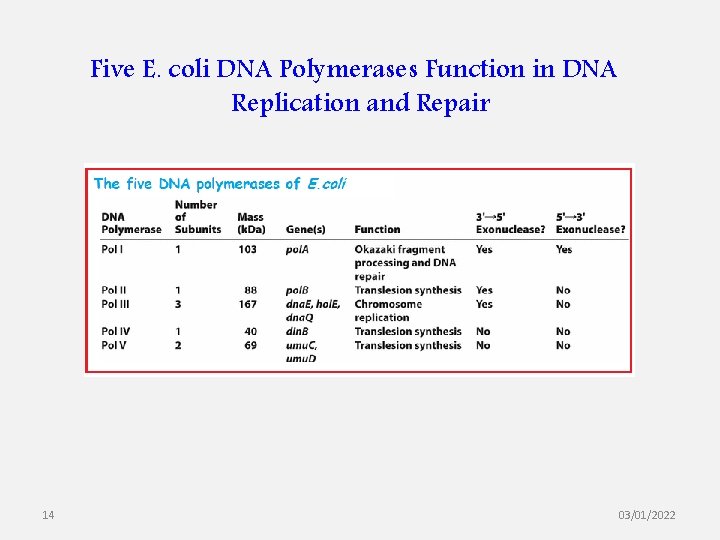
Five E. coli DNA Polymerases Function in DNA Replication and Repair 14 03/01/2022

ü DNA polymerases require a primed template and extend the 3′ terminus of the primer strand by reaction with d. NTPs. ü A d. NTP that correctly base-pairs to the template strand is incorporated into the primer strand with the release of pyrophosphate. Pyrophosphate is hydrolyzed by pyrophosphatase, which reduces the concentration of pyrophosphate in the cell and makes the reverse reaction extremely unlikely. ü DNA polymerases are inherently very accurate, and are made even more accurate by a proofreading 3′→ 5′ exonuclease. ü Pol I also contains a 5′→ 3′ exonuclease that degrades DNA while, at the same time, the polymerase synthesizes DNA, in the process of nick translation. ü E. coli has five DNA polymerases. Pol III is responsible for chromosome replication, and Pol I is used to remove RNA primers and 15 and fill in the resulting gaps with DNA. The other three (II, IV, 03/01/2022