DNA REPLICATION MAKING COPIES OVERVIEW DNA strands used





- Slides: 12

DNA REPLICATION MAKING COPIES

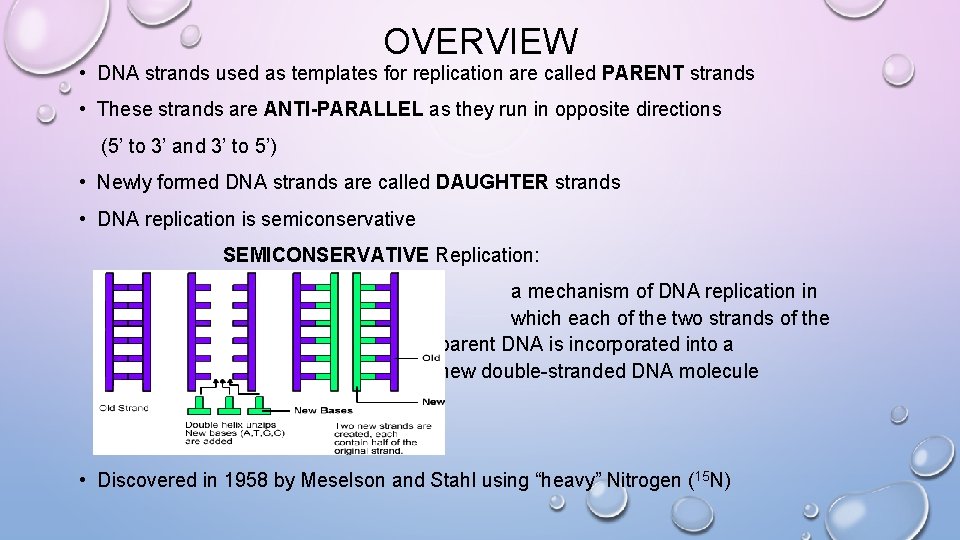
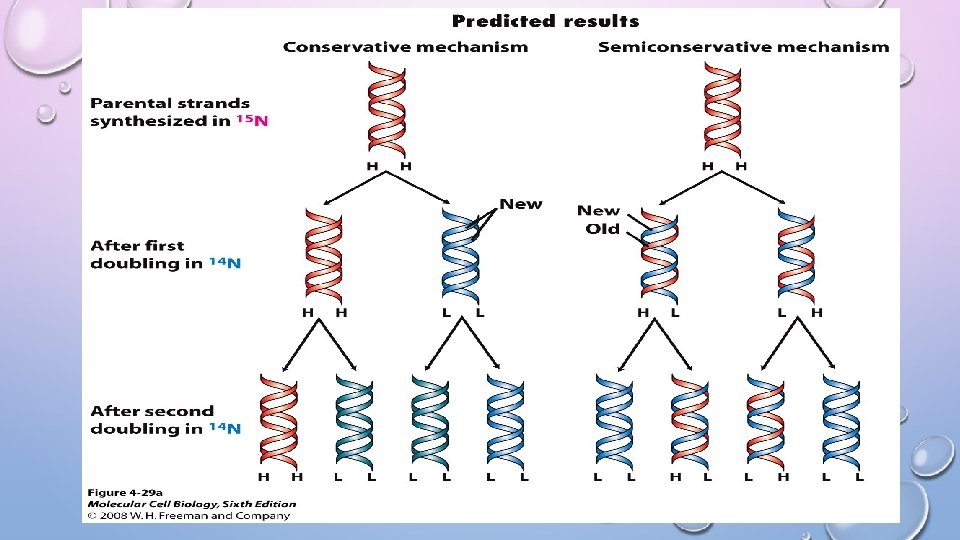
OVERVIEW • DNA strands used as templates for replication are called PARENT strands • These strands are ANTI-PARALLEL as they run in opposite directions (5’ to 3’ and 3’ to 5’) • Newly formed DNA strands are called DAUGHTER strands • DNA replication is semiconservative SEMICONSERVATIVE Replication: a mechanism of DNA replication in which each of the two strands of the parent DNA is incorporated into a new double-stranded DNA molecule • Discovered in 1958 by Meselson and Stahl using “heavy” Nitrogen (15 N)


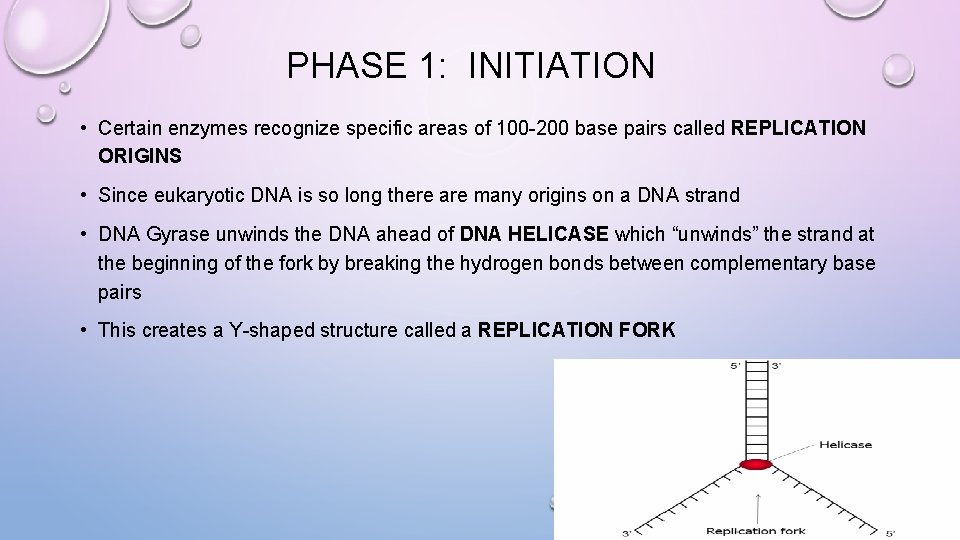
PHASE 1: INITIATION • Certain enzymes recognize specific areas of 100 -200 base pairs called REPLICATION ORIGINS • Since eukaryotic DNA is so long there are many origins on a DNA strand • DNA Gyrase unwinds the DNA ahead of DNA HELICASE which “unwinds” the strand at the beginning of the fork by breaking the hydrogen bonds between complementary base pairs • This creates a Y-shaped structure called a REPLICATION FORK

TENSION, ATTRACTION AND PROTEINS OH MY! • As the fork separates from the origin in both directions (REPLICATION BUBBLE), tension builds on the single strands • TOPOISOMERASES are enzymes that cut the strands to help unwind the strand relieve tension • Once this is accomplished the cut sections of the strand are reconnected by the same enzyme • The complimentary bases on the parent strands want to reform their Hydrogen bonds thus SINGLE-STRAND BINDING PROTEINS (SSBP) bind to each parent strand to prevent this from happening (prevent annealing) • These proteins are removed when the DNA is copied and reused when needed

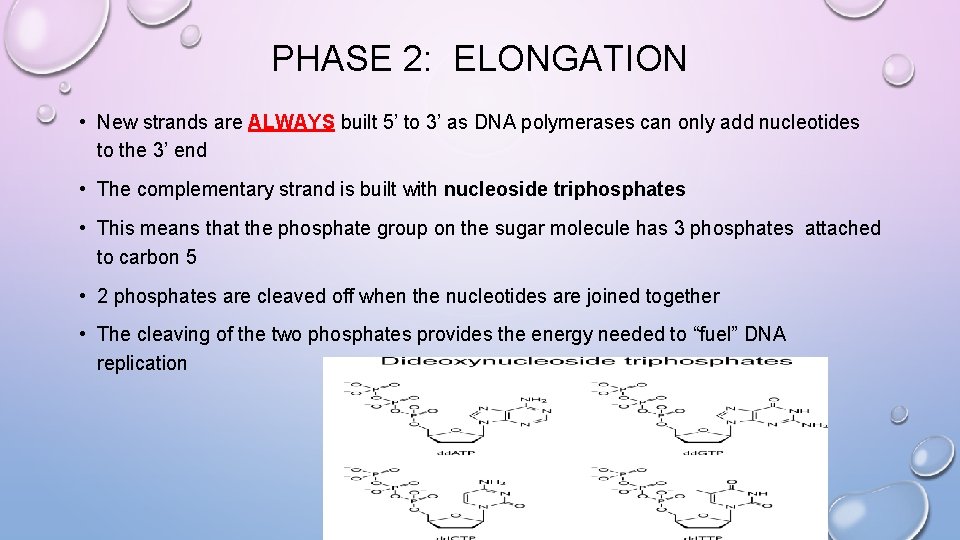
PHASE 2: ELONGATION • New strands are ALWAYS built 5’ to 3’ as DNA polymerases can only add nucleotides to the 3’ end • The complementary strand is built with nucleoside triphosphates • This means that the phosphate group on the sugar molecule has 3 phosphates attached to carbon 5 • 2 phosphates are cleaved off when the nucleotides are joined together • The cleaving of the two phosphates provides the energy needed to “fuel” DNA replication

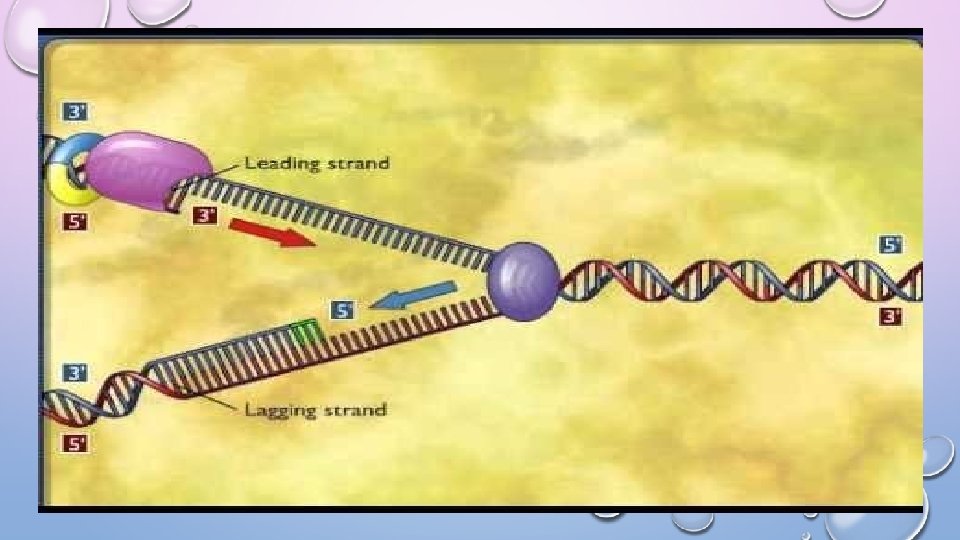
PHASE 2: THE LEADING STRAND • DNA polymerase can only attach nucleotides to a 3’ end so how does replication begin? • RNA PRIMERS are added by RNA PRIMASE at the beginning of the replication fork at the 3’ end of the template • RNA primase builds a complementary primer to this 3’ end and thus the new daughter strand is built in the 5’ to 3’ direction • Once the primer is in place DNA POLYMERASE III adds complementary nucleotides to the 3’ end of the daughter strand • This strand builds TOWARDS the replication fork and continues as the DNA unwinds thus ONLY ONE primer is required at each bubble. • This strand is called the LEADING STRAND

PHASE 2: THE LAGGING STRAND • The other parent strand has its 3’ end near the replication fork thus DNA polymerase must synthesize the new strand AWAY from the opening bubble (helicase) • when enough fork has opened RNA primase adds a primer near the fork • DNA polymerase III adds nucleotides until it reaches the end of the segment • When more of the fork opens RNA primase adds another primer and DNA polymerase III adds nucleotides until it reaches the last segment • This strand is built in small “pieces” called OKAZAKI FRAGMENTS • since this strand is not built in one continuous process it is called the LAGGING STRAND

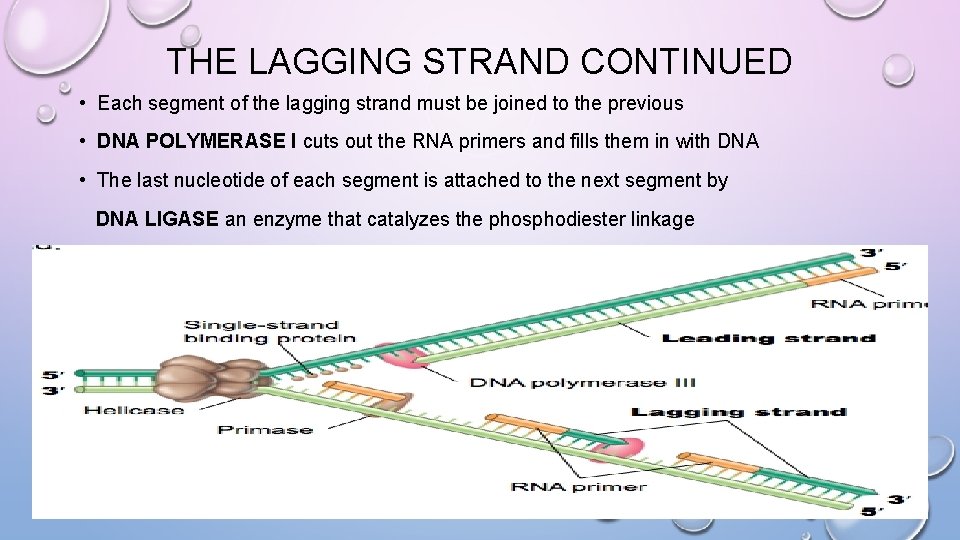
THE LAGGING STRAND CONTINUED • Each segment of the lagging strand must be joined to the previous • DNA POLYMERASE I cuts out the RNA primers and fills them in with DNA • The last nucleotide of each segment is attached to the next segment by DNA LIGASE an enzyme that catalyzes the phosphodiester linkage


PHASE 3: TERMINATION • DNA reforms its helical shape without the use of enzymes • At the end of OKAZAKI fragments there will be a small gap from the RNA primer • These unpaired DNA bases don’t mesh with the old strand, and fall off, leaving the new strand slightly shorter than the original • Humans lose approx. 100 base pairs per replication • Luckily the end of a chromosome there are telomeres (usually many G’s, in humans it is a repeat of TTAGGG)

PROOF READING • As DNA polymerase III moves along it proof reads the base pairs to ensure they are complementary • If there is a mismatch DNA polymerase III will not move forward, instead it will cut out the mismatch and replace it with the correct base pair • Leads to an error rate of 1 in 1 000 base pairs • A second complex comes through after DNA is made that contains proteins mainly DNA polymerase I, DNA polymerase II and DNA ligase • DNA POLYMERASE II moves slowly along the strand looks for shape distortions • Cleaves out incorrect strand replaces it with correct sequence • DNA polymerase II also used for repair of DNA damaged by environmental causes