DNA Replication Campbell Chap 16 pg 284 Inquiry
DNA Replication Campbell- Chap 16 pg 284 Inquiry Chap 25
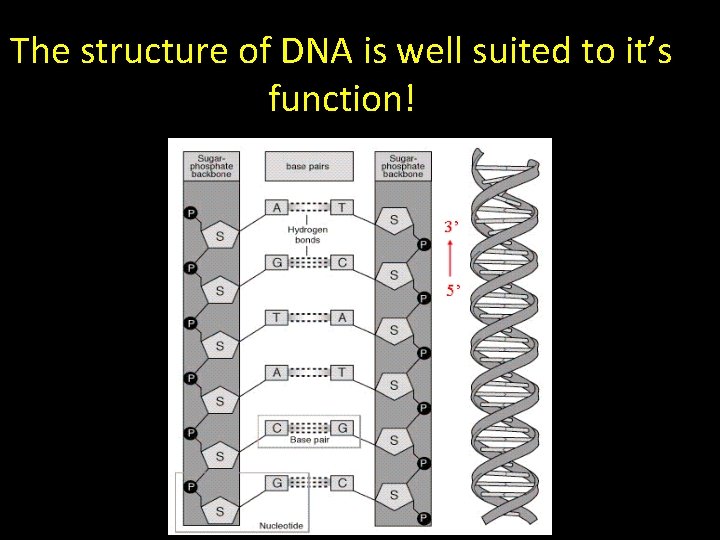
The structure of DNA is well suited to it’s function!
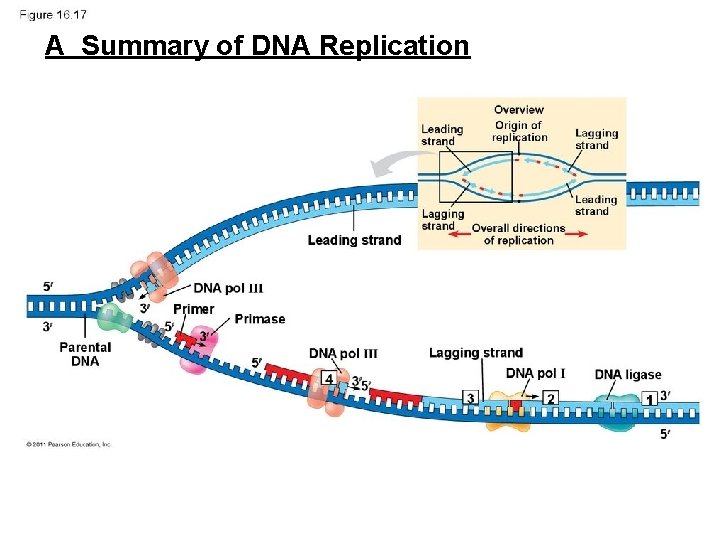
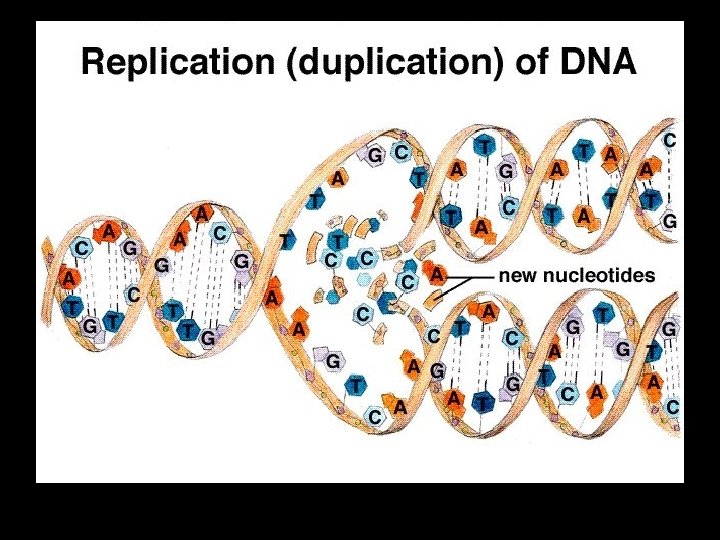
AA Summary of DNA Replication
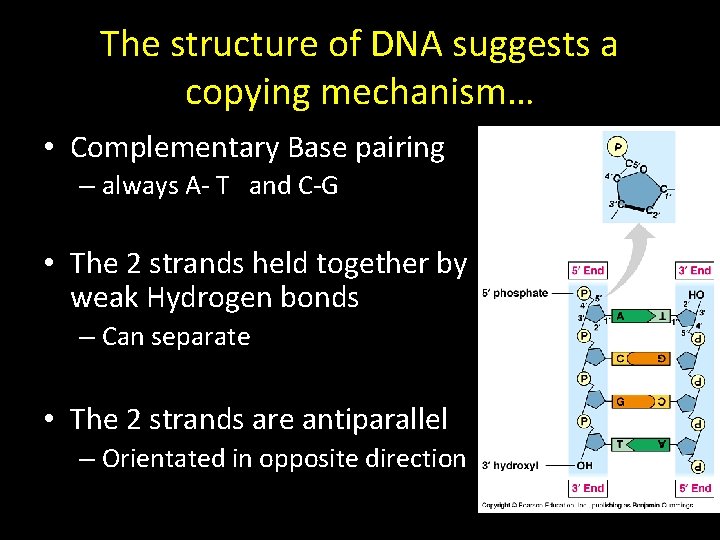
The structure of DNA suggests a copying mechanism… • Complementary Base pairing – always A- T and C-G • The 2 strands held together by weak Hydrogen bonds – Can separate • The 2 strands are antiparallel – Orientated in opposite direction

• Many enzymes & other proteins are also needed to carry out replication!
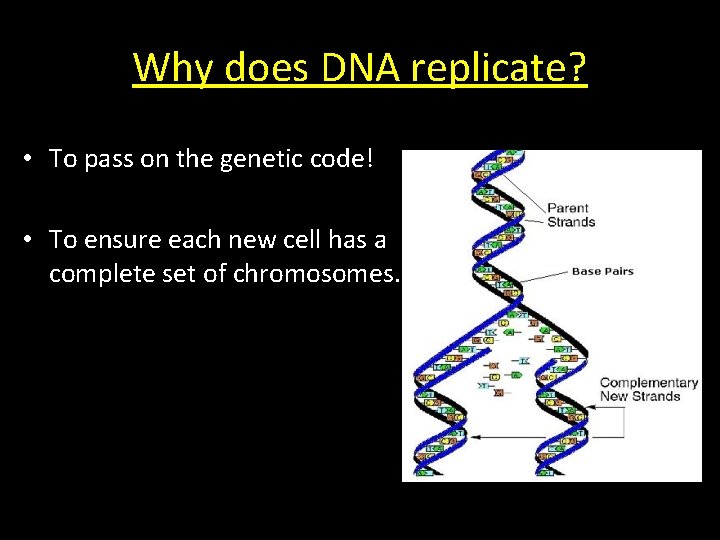
Why does DNA replicate? • To pass on the genetic code! • To ensure each new cell has a complete set of chromosomes.
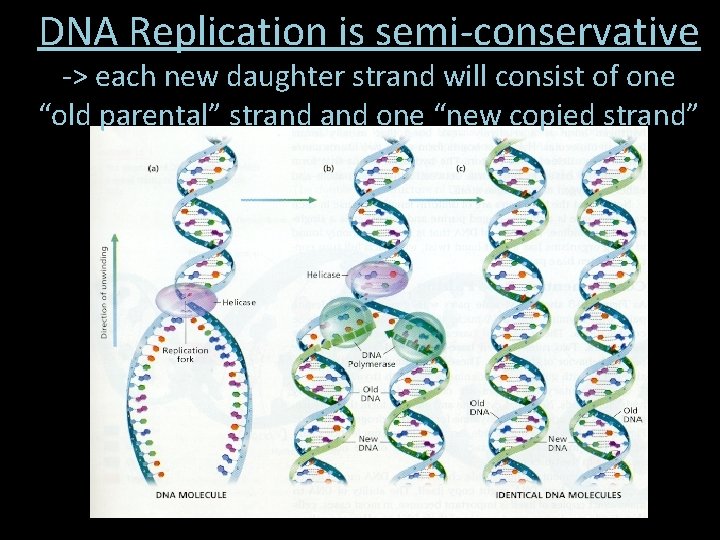
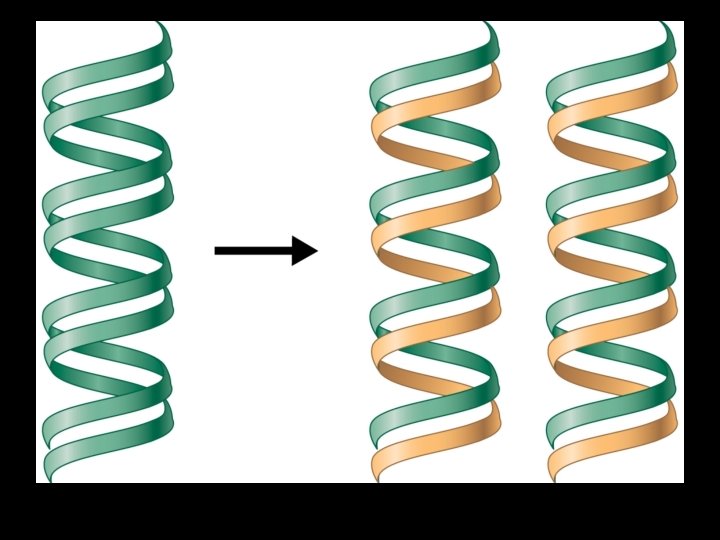
DNA Replication is semi-conservative -> each new daughter strand will consist of one “old parental” strand one “new copied strand”

Helpful Replication Clips • 3 D Animation: https: //www. youtube. com/watch? v=TNKWgc. FPHq w • Animation: https: //www. youtube. com/watch? v=Vpm. T 7 Lw_4 v 0 • Ameoba Sisters: https: //www. youtube. com/watch? v=5 q. Srmei. Wsuc • Crash Course: https: //www. youtube. com/watch? v=8 k. K 2 zwj. RV 0 M • The immortal cells of Henriette Lacks: https: //www. youtube. com/watch? v=22 l. Gb. AVWhr
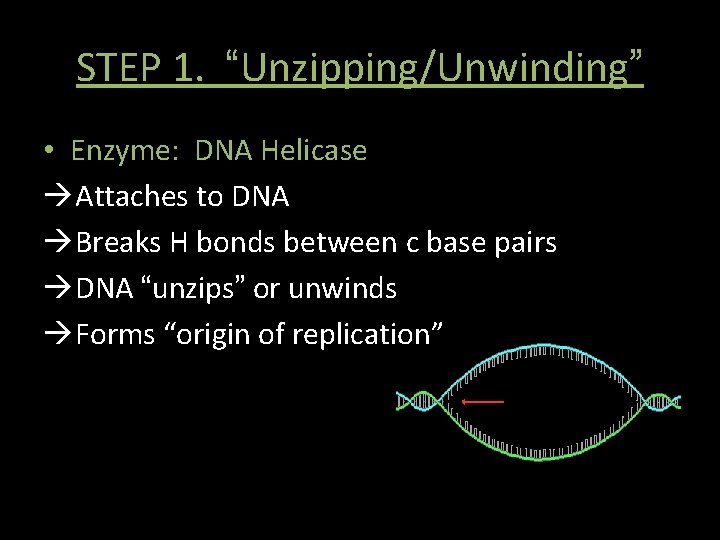
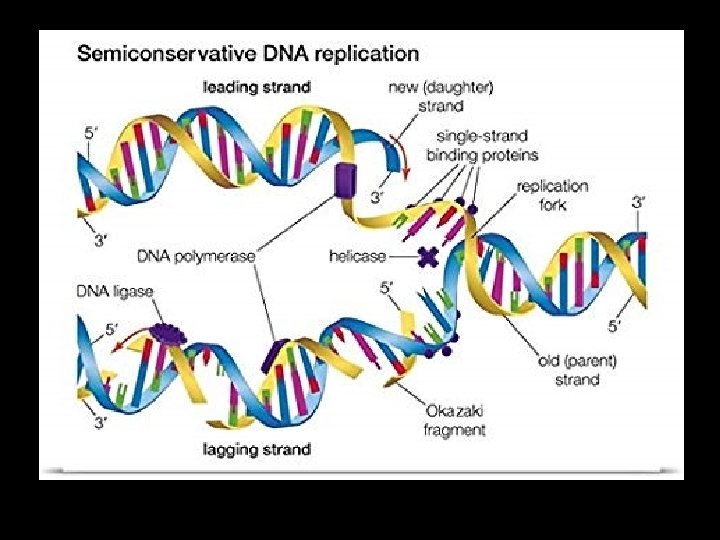
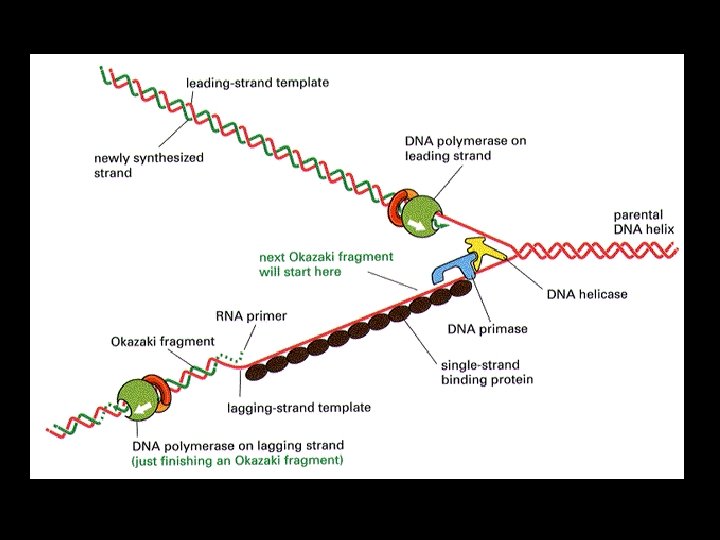
STEP 1. “Unzipping/Unwinding” • Enzyme: DNA Helicase Attaches to DNA Breaks H bonds between c base pairs DNA “unzips” or unwinds Forms “origin of replication”
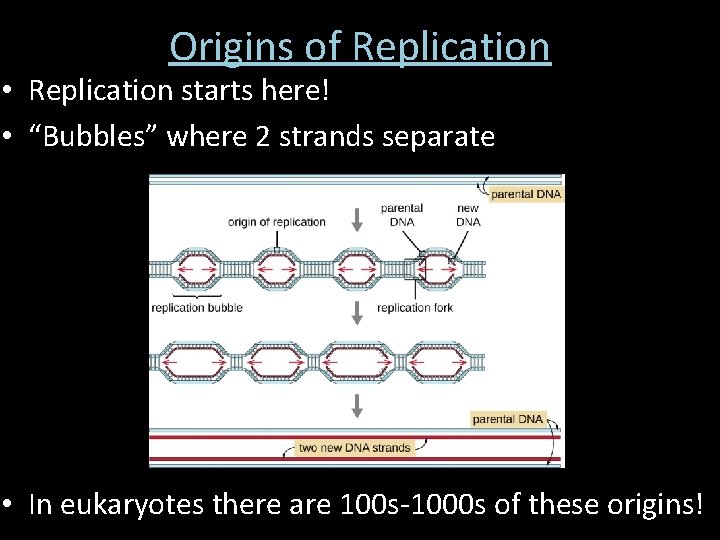
Origins of Replication • Replication starts here! • “Bubbles” where 2 strands separate • In eukaryotes there are 100 s-1000 s of these origins!
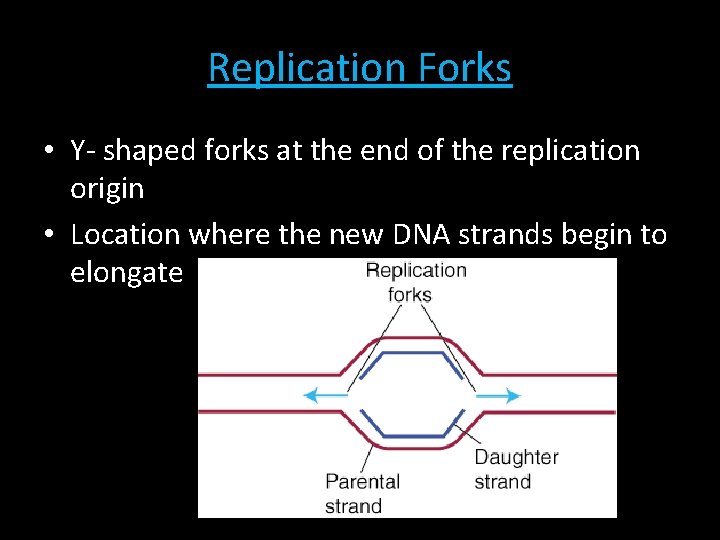
Replication Forks • Y- shaped forks at the end of the replication origin • Location where the new DNA strands begin to elongate

How does the anti-parallel nature of the strands affect replication? • The new DNA strand can ONLY elongate in the 5’ 3’ direction • Nucleotides will ONLY get added to the free 3’ end
2. Elongating- Complementary base pairing • DNA polymerases catalyse this step • A RNA primer is required to start – Primase enzymes join RNA nucleotides to create it
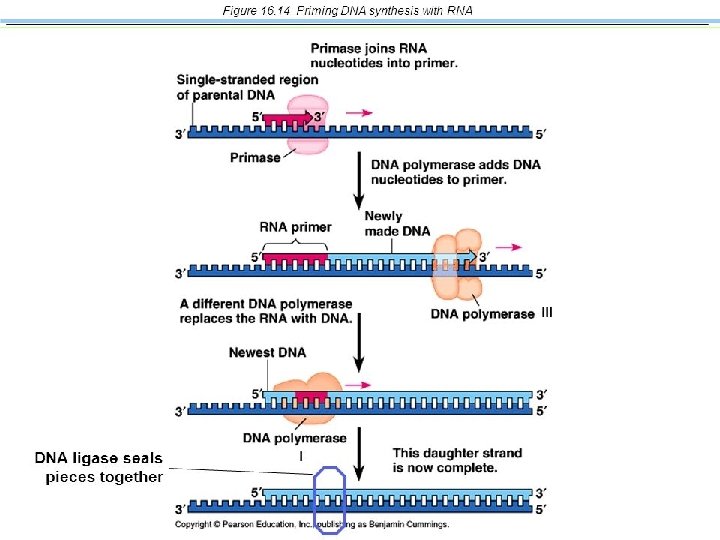
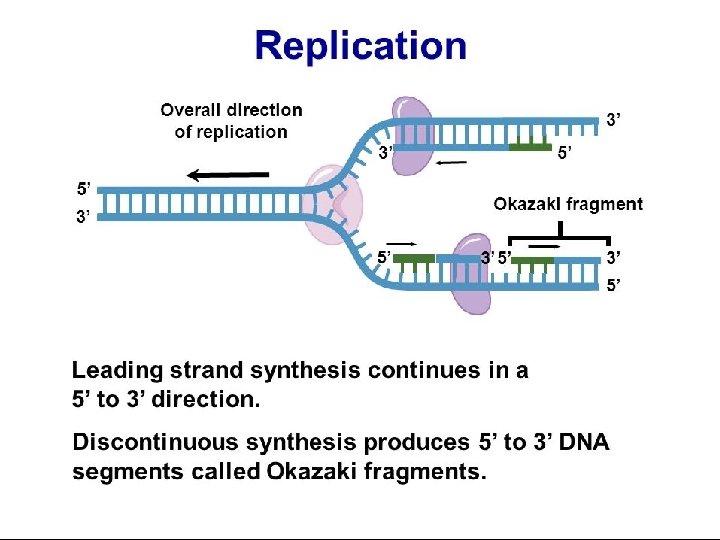
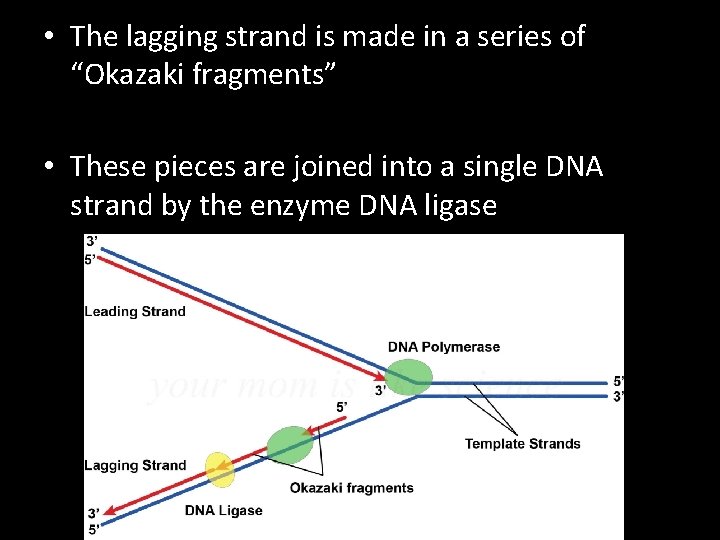
• The lagging strand is made in a series of “Okazaki fragments” • These pieces are joined into a single DNA strand by the enzyme DNA ligase
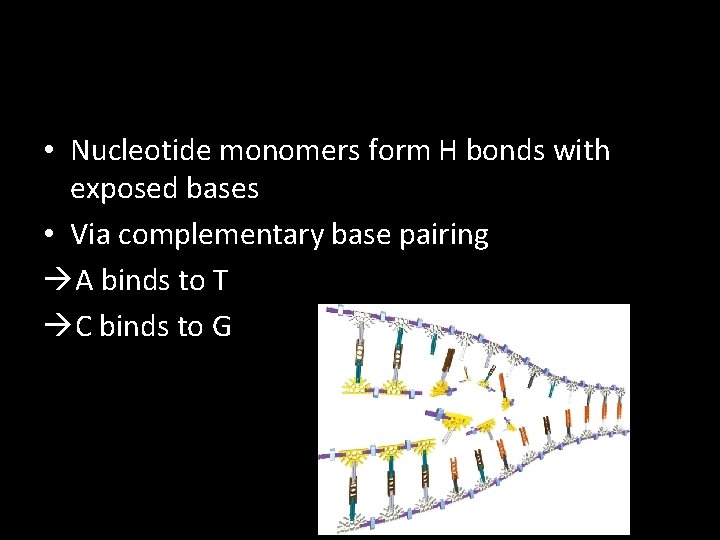
• Nucleotide monomers form H bonds with exposed bases • Via complementary base pairing A binds to T C binds to G

3. Joining of adjacent nucleotides • Enzyme: DNA Polymerase This step creates the new sugar-phosphate backbone formation of sugar-phosphate covalent bonds (dehydration synthesis) between each nucleotide

4. Proofreading of new DNA • Mismatch repair – DNA polymerase can also fix mistakes made during replication – Any incorrectly paired nucleotide is removed • Excision repair – Errors “cut out” by nuclease – DNA polymerase fills the gap – Ligase seals the nick • There are many DNA repair enzymes!

The ends of DNA pose a problem… • DNA polymerasae can only add nucleotides to the 3’ end… • There’s no way to complete the 5’ ends of the daughter strands – Over time replicated strands get shorter & shorter • Solution: Telomeres – Typically, they are a repeated segment of TTAGGG that doesn’t code for any genes at the end of DNA

Replication Animation • https: //www. youtube. com/watch? v=y. Dk. SWd _Zbb. E
- Slides: 25