DNA replication By Dr Ahmed Okasha Semiconservative replication
DNA replication By Dr. Ahmed Okasha

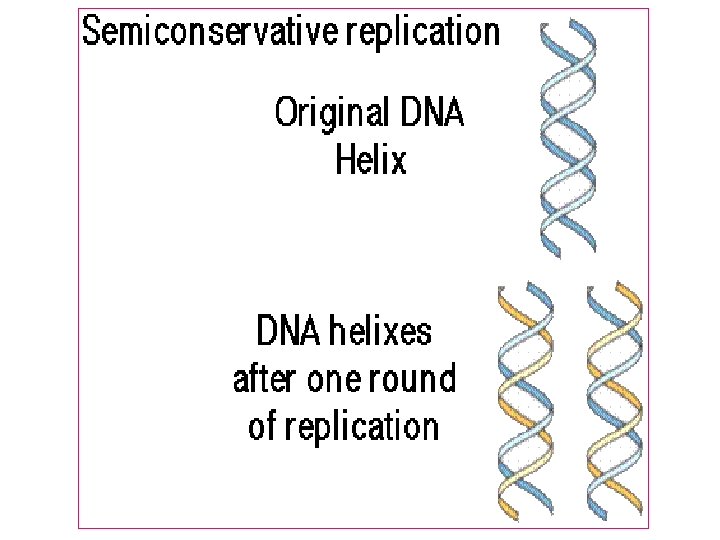
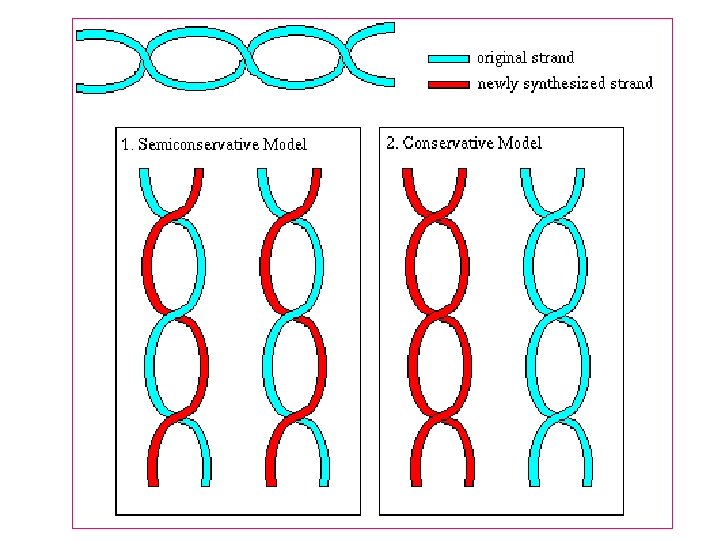
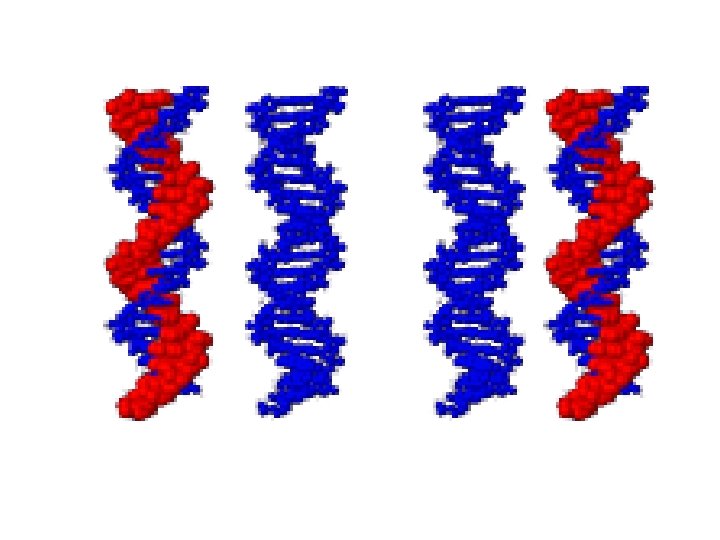
Semiconservative replication 1 -Separation of DNA strand occur 2 -The new strand synthesized that are complementary to parent strand 3 -The new DNA contain 2 strands ne from parents and the other is new strand

Basic requirement for DNA replication 1 - Substrate 4 nucleotide of DNA (ATP, GTP, CTP, TTP) must present 2 - Template (parent strand) Parent strand are required to add complementary nucleotide on the new strand
3 - RNA primer Present in the first new strand its function is to receive the first new nucleotide that added on the new strand 4 - Enzyme : DNA polymerase , Pol I, III

Step of replication in prokaryotic (E. coli)
Origin of replication 1 -Replication start at particular DNA sequence called origin (ori) 2 - Dna A & C protein bind to strand produce separation of 2 strand

2 - Formation of replication fork Replication fork represent the unwinding the DNA strand by the effect of helicase (Dna B) made broken in hydrogen bond lead to separation in helix This need 2 ATP for each base pair broken

Helicaes helped by 2 enzyme Topoisomerase Prevent super coil (super twist) that if occur lead to prevent replication
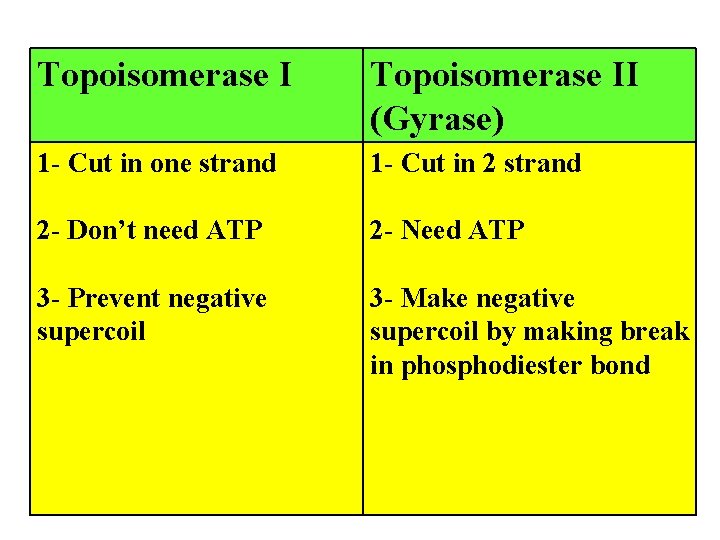
Topoisomerase II (Gyrase) 1 - Cut in one strand 1 - Cut in 2 strand 2 - Don’t need ATP 2 - Need ATP 3 - Prevent negative supercoil 3 - Make negative supercoil by making break in phosphodiester bond

Single stranded binding protein (SSBP) Bind to single stranded DNA prevent it from reunion

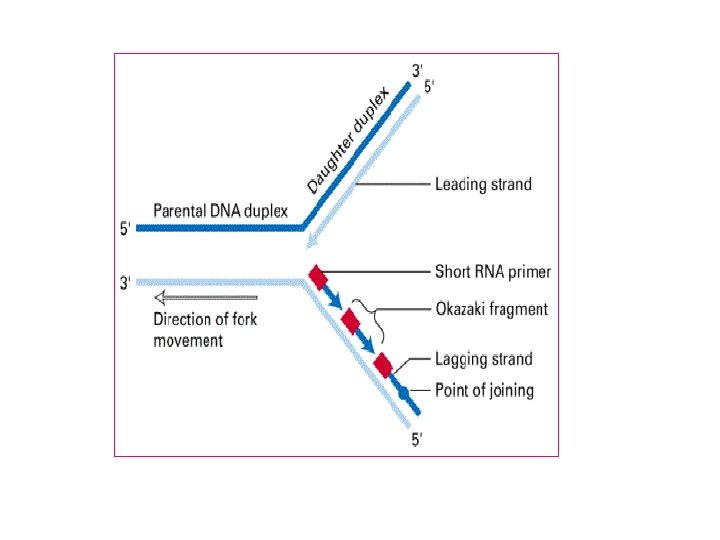
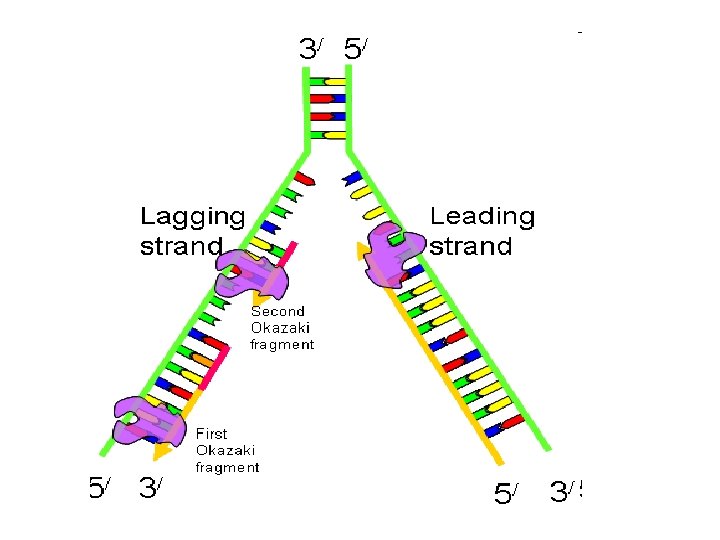
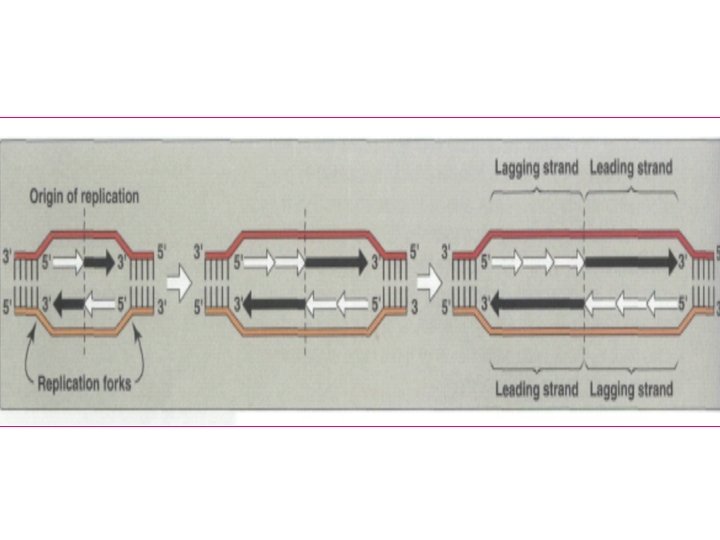
Formation of new complementary strand Always new DNA strand grow from strand Leading AThis strand grow from 5 to 3 indirection of origin of replication This grow continuously and made by

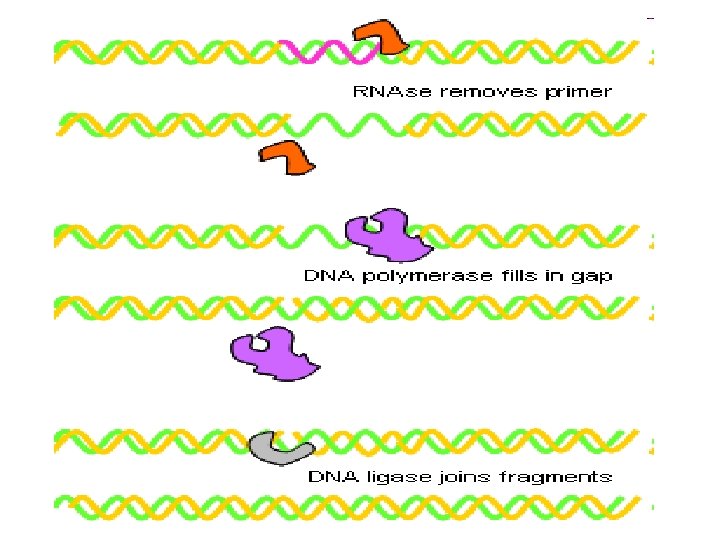
B) Okazaki fragment (lagging strand) This strand grow from 5 to 3 direction in osite of origin of replication This grow discontinuously and made by The gap (space) filled by DNA Pol I and the space is ligated By DNA ligase
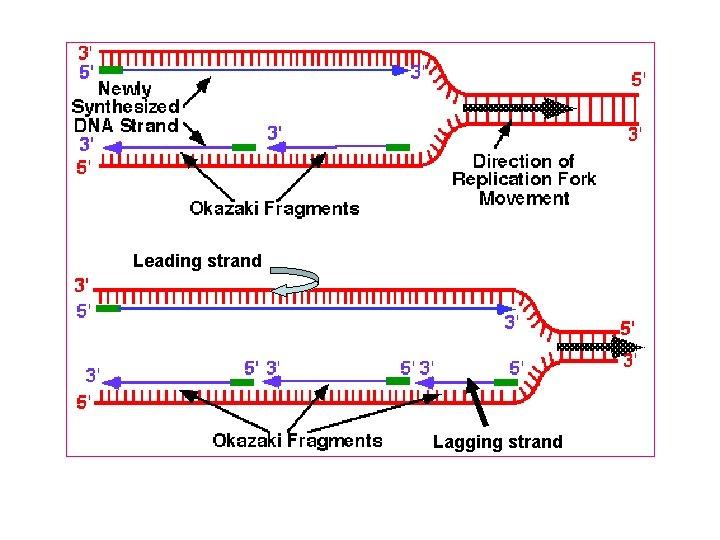
Leading strand Lagging strand
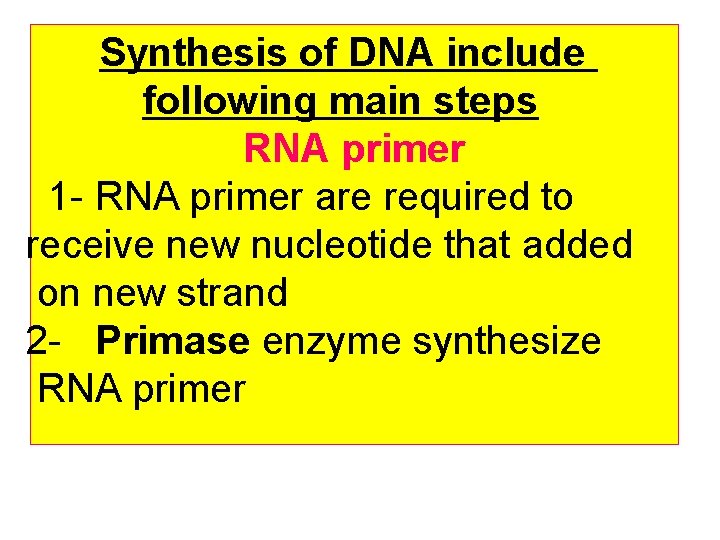
Synthesis of DNA include following main steps RNA primer 1 - RNA primer are required to receive new nucleotide that added on new strand 2 - Primase enzyme synthesize RNA primer
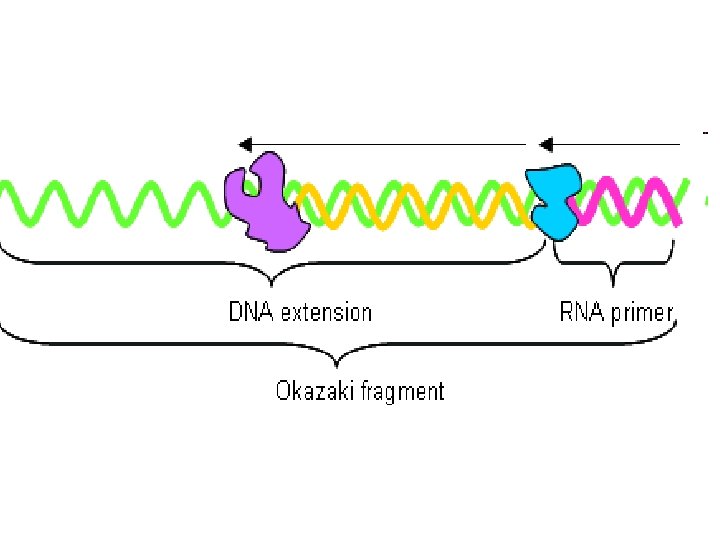

Pol I remove primer after termination of replication There are 1 primer on leading strand many primer in lagging strand Prof reading effect of DNA Pol III Prof reading means ability to remove any wrong nucleotide added during synthesis as pairing A=G must be A=T

DNA Pol II Has role in repair of DNA NB. Replication of DNA has high fidelity means any error during synthesis is corrected
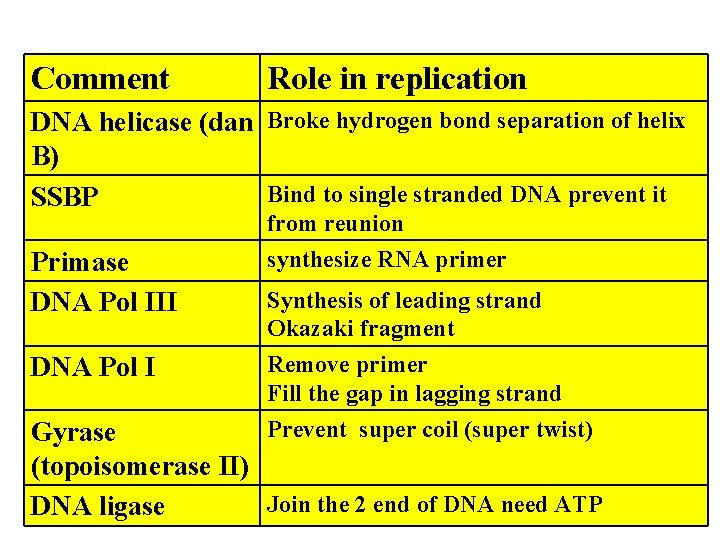
Comment Role in replication DNA helicase (dan Broke hydrogen bond separation of helix B) Bind to single stranded DNA prevent it SSBP from reunion Primase DNA Pol III DNA Pol I synthesize RNA primer Synthesis of leading strand Okazaki fragment Remove primer Fill the gap in lagging strand Prevent super coil (super twist) Gyrase (topoisomerase II) Join the 2 end of DNA need ATP DNA ligase
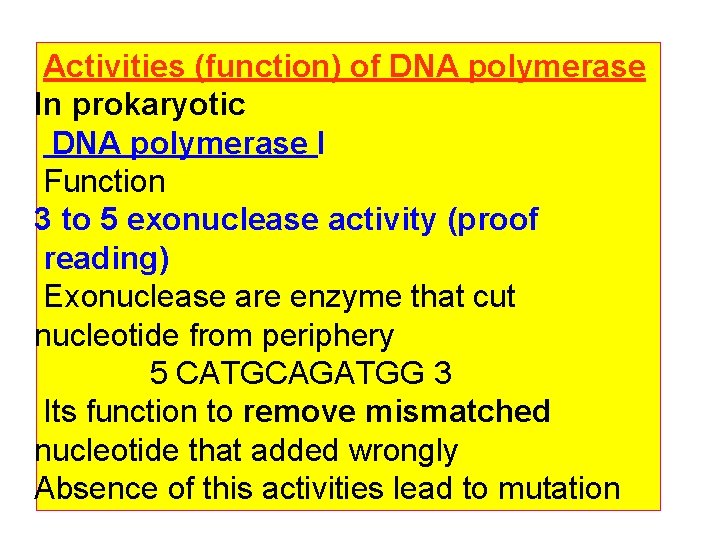
Activities (function) of DNA polymerase In prokaryotic DNA polymerase I Function 3 to 5 exonuclease activity (proof reading) Exonuclease are enzyme that cut nucleotide from periphery 5 CATGCAGATGG 3 Its function to remove mismatched nucleotide that added wrongly Absence of this activities lead to mutation

5 to 3 exonuclease activity ) repair 5 CATGCAGATGG 3 1 -Also has role in removal RNA primer after replication 2 - Has role in repair damaged DNA

DNA polymerase II Has role in repair of DNA polymerase III Synthesis of leading strand , Okazaki fragment Eukaryotic DNA polymerase Types α, β, γ, δ, ε
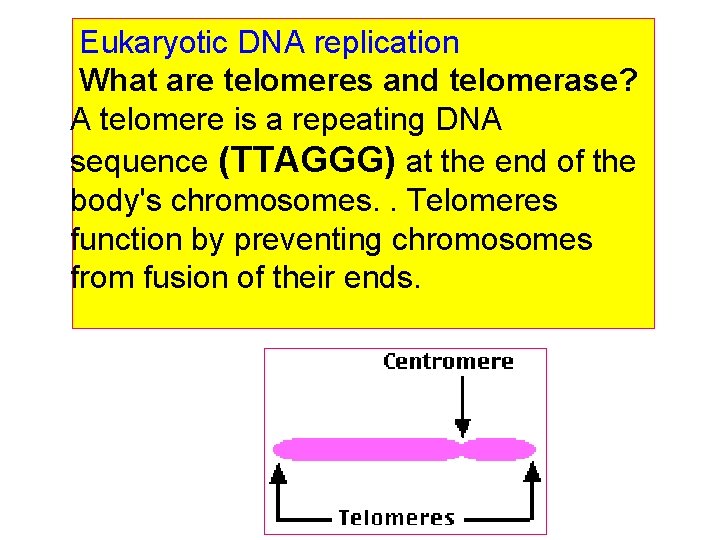
Eukaryotic DNA replication What are telomeres and telomerase? A telomere is a repeating DNA sequence (TTAGGG) at the end of the body's chromosomes. . Telomeres function by preventing chromosomes from fusion of their ends.

However, each time a cell divides, some of the telomere is lost (usually 25 -200 base pairs per division). When the telomere becomes too short, the chromosome reaches a "critical length" and can no longer replicate. This means that a cell becomes "old“ senescent and dies
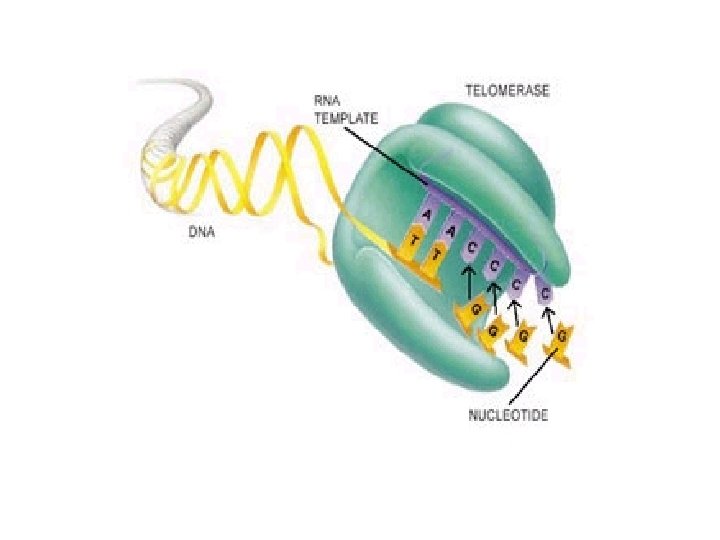
Telomerase, is an enzyme made of protein and RNA subunits that elongates chromosomes by adding TTAGGG sequences to the end chromosomes.
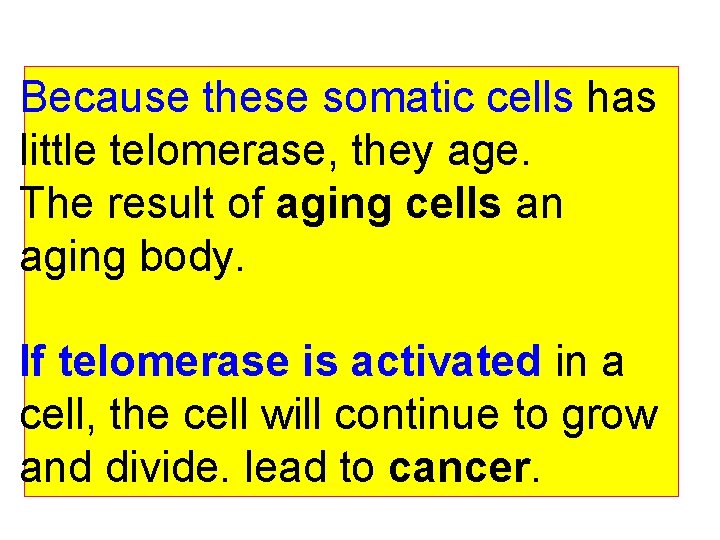
Because these somatic cells has little telomerase, they age. The result of aging cells an aging body. If telomerase is activated in a cell, the cell will continue to grow and divide. lead to cancer.
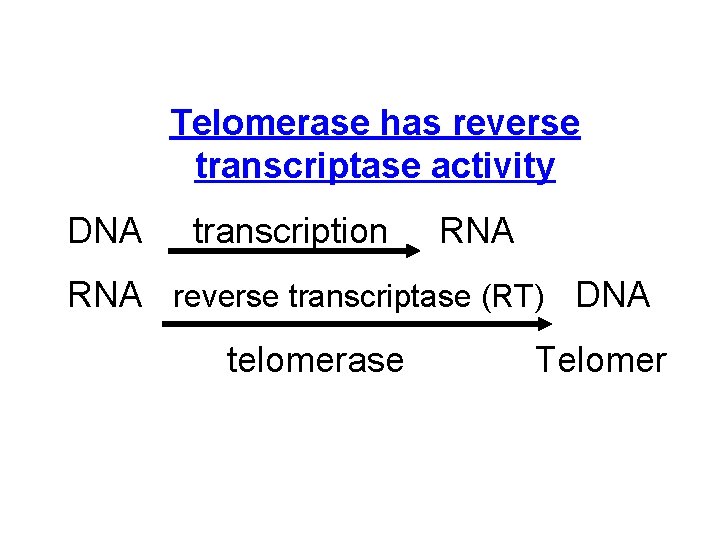
Telomerase has reverse transcriptase activity DNA transcription RNA reverse transcriptase (RT) DNA telomerase Telomer
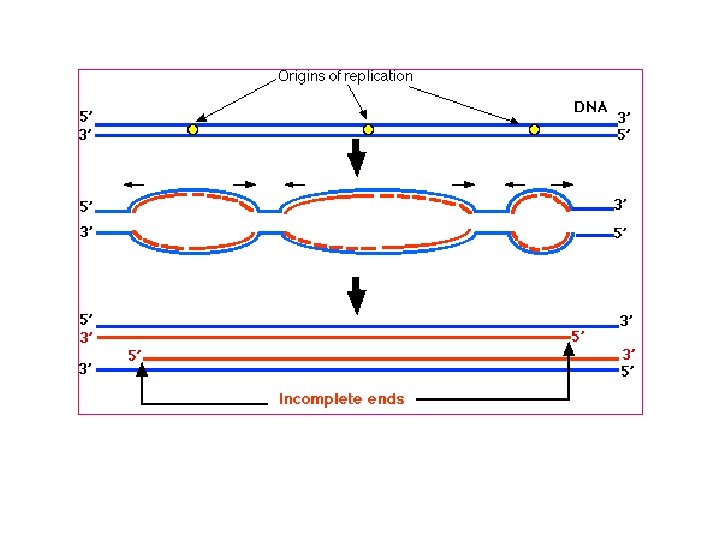
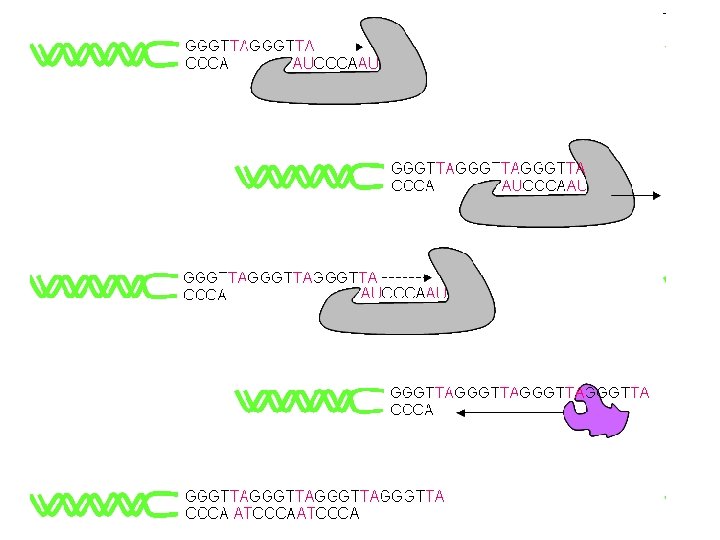
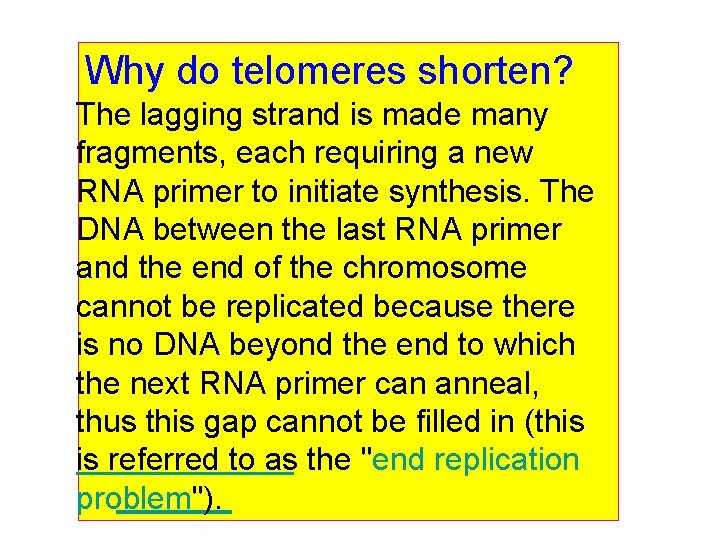
Why do telomeres shorten? The lagging strand is made many fragments, each requiring a new RNA primer to initiate synthesis. The DNA between the last RNA primer and the end of the chromosome cannot be replicated because there is no DNA beyond the end to which the next RNA primer can anneal, thus this gap cannot be filled in (this is referred to as the "end replication problem").

Since one strand cannot copy its end, telomere shortening occurs during progressive cell divisions. If you can stop the shortening of telomeres will this prevent cellular aging cancer cells show no net loss of average telomere length with cell division
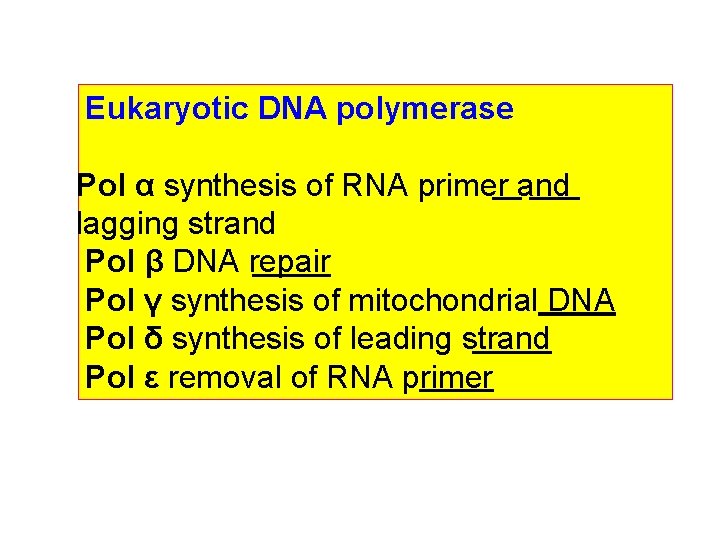
Eukaryotic DNA polymerase Pol α synthesis of RNA primer and lagging strand Pol β DNA repair Pol γ synthesis of mitochondrial DNA Pol δ synthesis of leading strand Pol ε removal of RNA primer

Difference between Eukaryotic and prokaryotic DNA replication
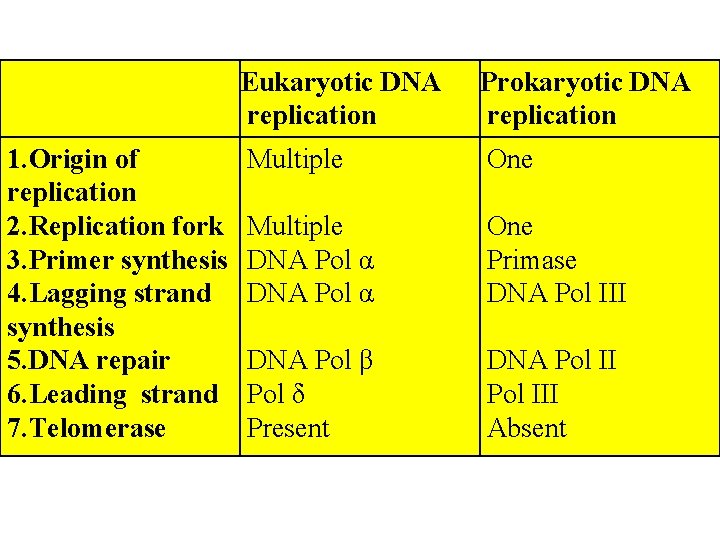
1. Origin of replication 2. Replication fork 3. Primer synthesis 4. Lagging strand synthesis 5. DNA repair 6. Leading strand 7. Telomerase Eukaryotic DNA replication Prokaryotic DNA replication Multiple One Multiple DNA Pol α One Primase DNA Pol III DNA Pol β Pol δ Present DNA Pol III Absent

- Slides: 43