DNA repair Mutacin y reparacin del DNA Why



















- Slides: 45

DNA repair Mutación y reparación del DNA

Why repair DNA? Replication error • 1. Errors in DNA replication • 2. Endogenous DNA damage and mutagens • 3. Environmental insults to DNA • 4. Un-repaired damage leads to: • -mistakes in RNA/protein synthesis • -inherited as genetic alteration-a mutation • -death OH. d. UTP UV hn H+ m. C TT P/P 8 -oxo. G U Depurination All of these events are rare, but the number of bp in each nucleus is very large so the total frequency is significant.

Types of DNA damage • Base Loss • Base modification & Deamination • Chemical Modification • Photodamage • Inter-strand crosslinks • DNA-protein crosslinks • Strand breakage

Example of the product of a very small and a very large number yielding a significant effect • Number of bp in the nucleus of a human cell=~3 109. • Rate of breakage of purine glycosidic bonds in neutral solution predicts =~104 depurinations/day/cell. • 1013 cells/human = 1017 depurinations/day/organism! • If these are not repaired, it would lead to massive errors in the synthesis of proteins. • Mutations in the germ line would transmitted to offspring, leading to genetic disease.

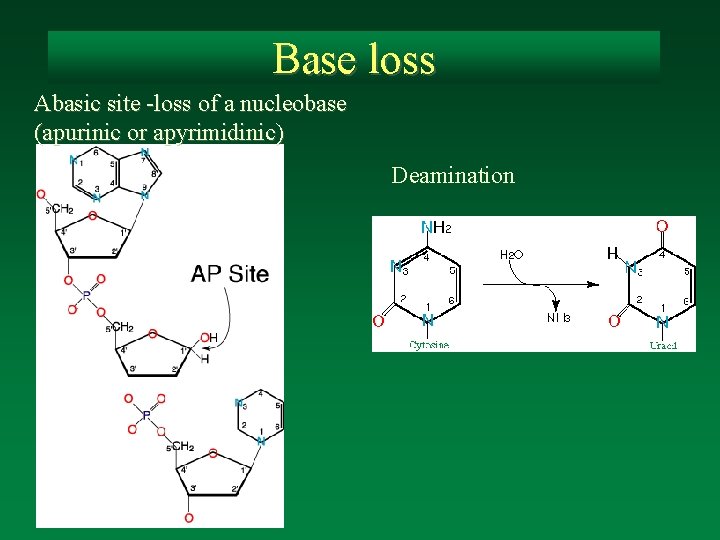
Base loss Abasic site -loss of a nucleobase (apurinic or apyrimidinic) Deamination

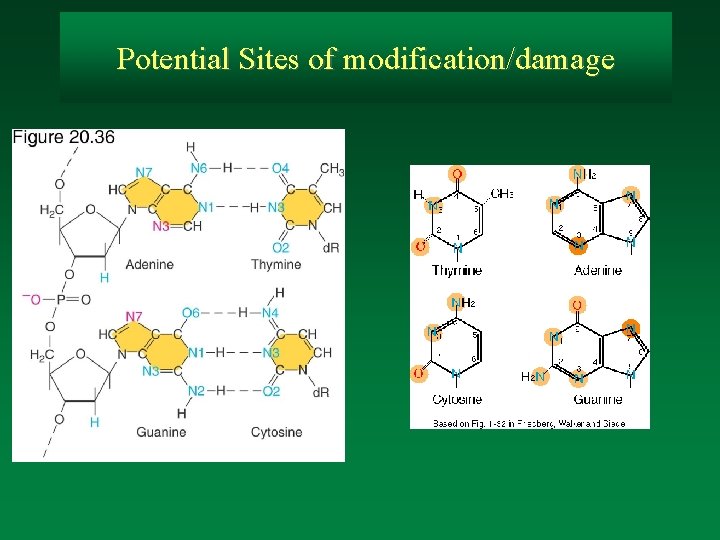
Potential Sites of modification/damage

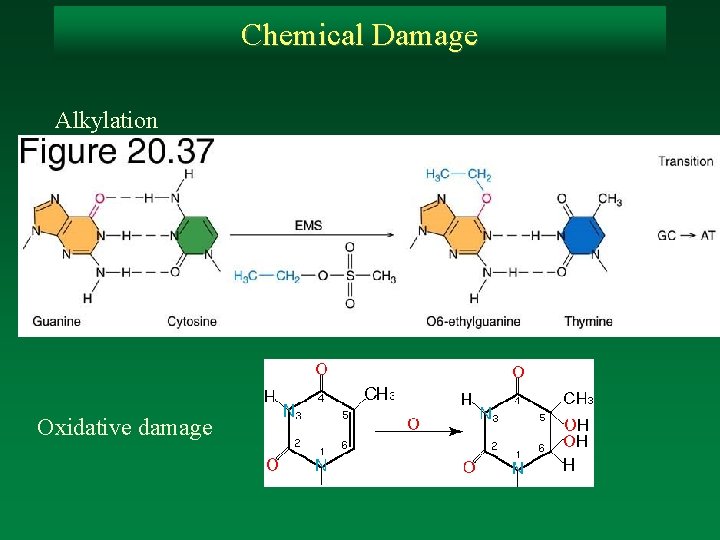
Chemical Damage Alkylation Oxidative damage

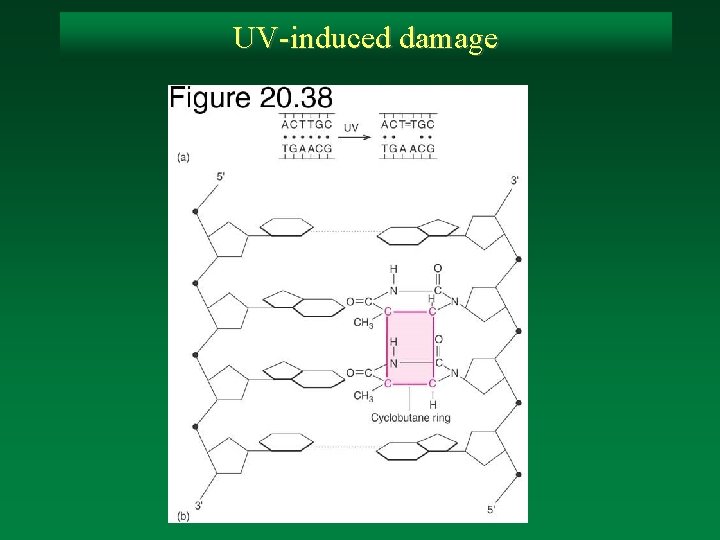
UV-induced damage

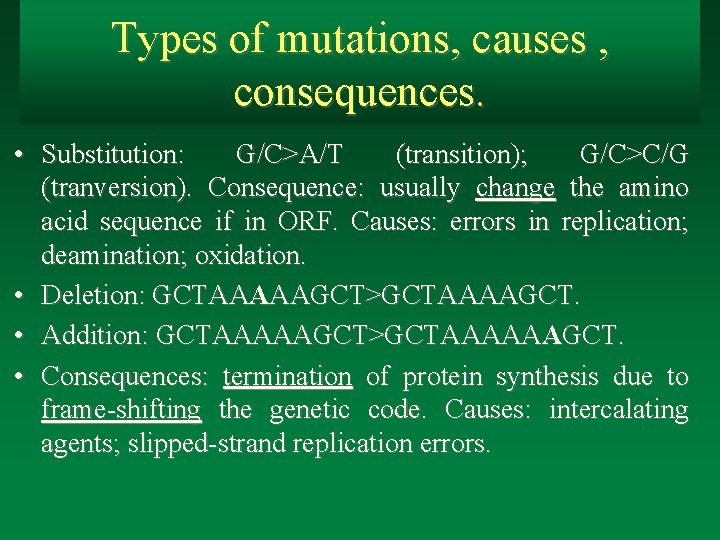
Types of mutations, causes , consequences. • Substitution: G/C>A/T (transition); G/C>C/G (tranversion). Consequence: usually change the amino acid sequence if in ORF. Causes: errors in replication; deamination; oxidation. • Deletion: GCTAAAAAGCT>GCTAAAAGCT. • Addition: GCTAAAAAGCT>GCTAAAAAAGCT. • Consequences: termination of protein synthesis due to frame-shifting the genetic code. Causes: intercalating agents; slipped-strand replication errors.

Consequences of mutations (cont) Accumulation of mutations: • Cancer, (Loeb’s hypothesis): cancer is a genetic “disease” caused by an elevated mutation rate-as by an error-prone polymerase or faulty repair machinery. “Two-hit” model. Somatic versus germ line mutations. • Aging (error catastrophe hypothesis); failure of normal cell death (apoptosis) due to accumulation of mutations in genes responsible for the normal operation of these processes. • Single mutations and genetic disease. How many genes, when inactivated, would cause a “disease”? • Multiple polymorphisms (remember SNPs) and predisposition to susceptibility to endogenous agents (oxidizing agents) and environmental insults.

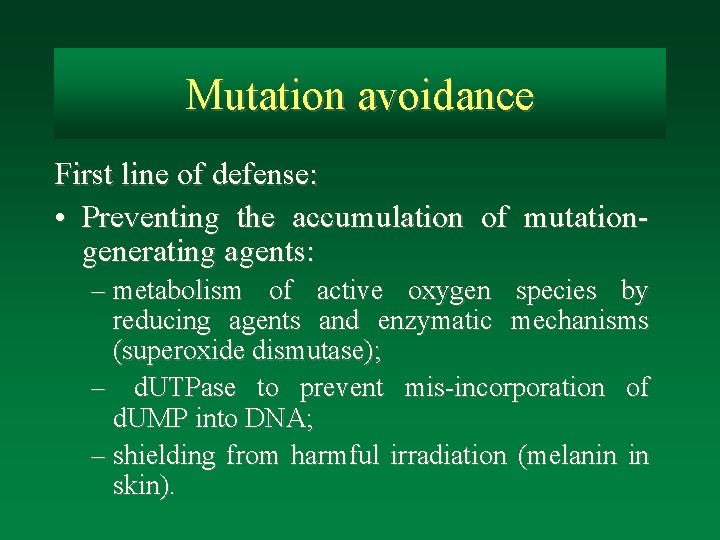
Mutation avoidance First line of defense: • Preventing the accumulation of mutationgenerating agents: – metabolism of active oxygen species by reducing agents and enzymatic mechanisms (superoxide dismutase); – d. UTPase to prevent mis-incorporation of d. UMP into DNA; – shielding from harmful irradiation (melanin in skin).

Types of Damage Repair • Photolyase • De-alkylation proteins (not catalytic) • Base Excision Repair • Nucleotide Excision Repair (GG and TC) • • • Recombination Repair Error-prone Repair Double strand Break Repair (if time permits)

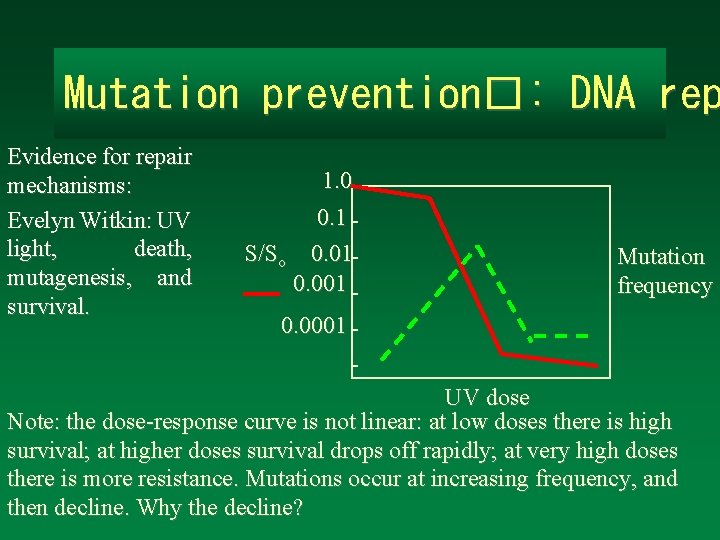
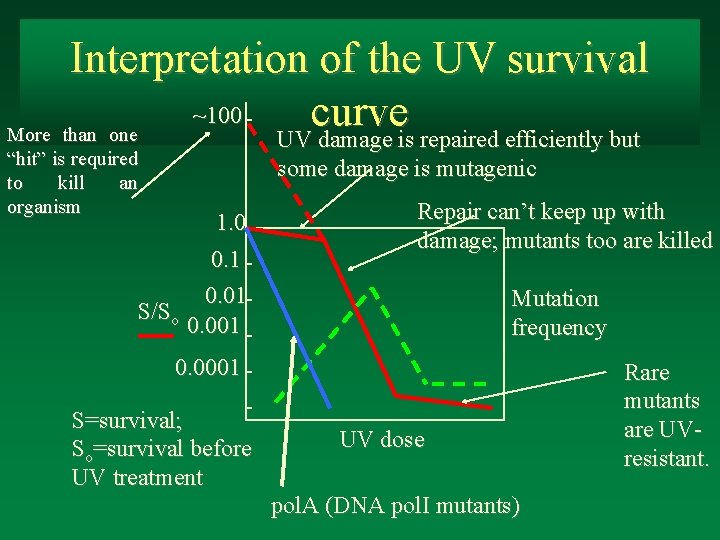
Mutation prevention�: DNA rep Evidence for repair mechanisms: Evelyn Witkin: UV light, death, mutagenesis, and survival. 1. 0 0. 1 S/So 0. 01 0. 001 Mutation frequency 0. 0001 UV dose Note: the dose-response curve is not linear: at low doses there is high survival; at higher doses survival drops off rapidly; at very high doses there is more resistance. Mutations occur at increasing frequency, and then decline. Why the decline?

Interpretation of the UV survival ~100 curve More than one UV damage is repaired efficiently but “hit” is required to kill an organism some damage is mutagenic 1. 0 0. 1 0. 01 S/So 0. 001 Repair can’t keep up with damage; mutants too are killed Mutation frequency 0. 0001 S=survival; So=survival before UV treatment UV dose pol. A (DNA pol. I mutants) Rare mutants are UVresistant.

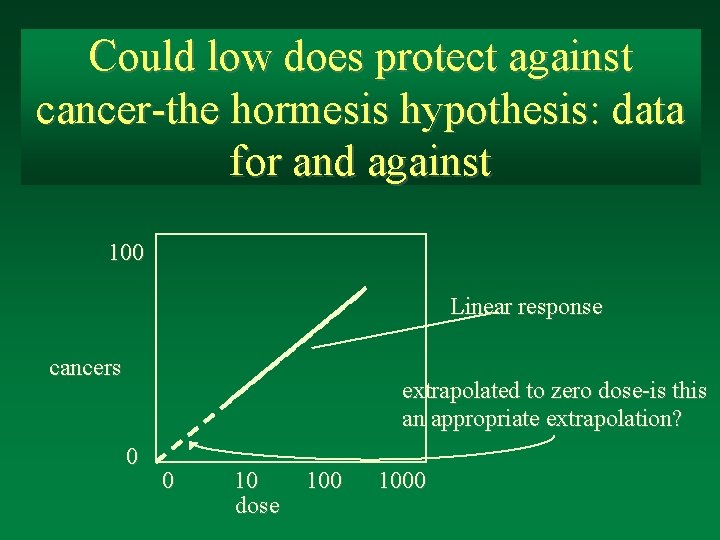
Could low does protect against cancer-the hormesis hypothesis: data for and against 100 Linear response cancers extrapolated to zero dose-is this an appropriate extrapolation? 0 0 10 dose 1000

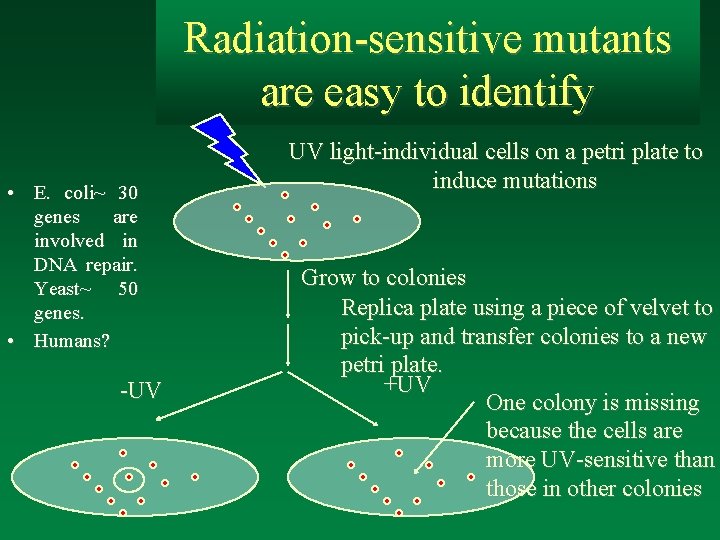
Radiation-sensitive mutants are easy to identify • E. coli~ 30 genes are involved in DNA repair. Yeast~ 50 genes. • Humans? -UV UV light-individual cells on a petri plate to induce mutations Grow to colonies Replica plate using a piece of velvet to pick-up and transfer colonies to a new petri plate. +UV One colony is missing because the cells are more UV-sensitive than those in other colonies

Redundancy of repair mechanisms • • 1. Proof-reading or editing by DNA polymerase 2. Direct reversal of damage. 3. Base excision repair 4. Nucleotide excision repair* 5. Mismatch repair. 6. Error-prone (SOS) repair* 7. Recombination repair * Induced by DNA damaging agents

Editing: Frequency of errors in replication depends on the polymerase

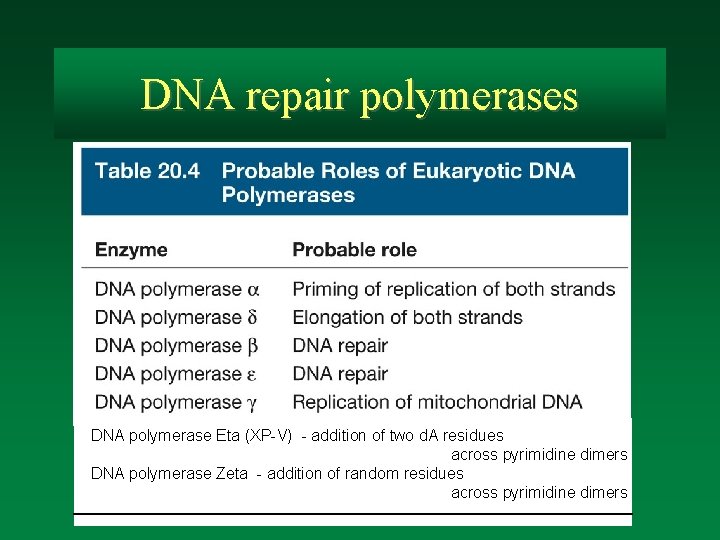
DNA repair polymerases DNA polymerase Eta (XP-V) - addition of two d. A residues across pyrimidine dimers DNA polymerase Zeta - addition of random residues across pyrimidine dimers

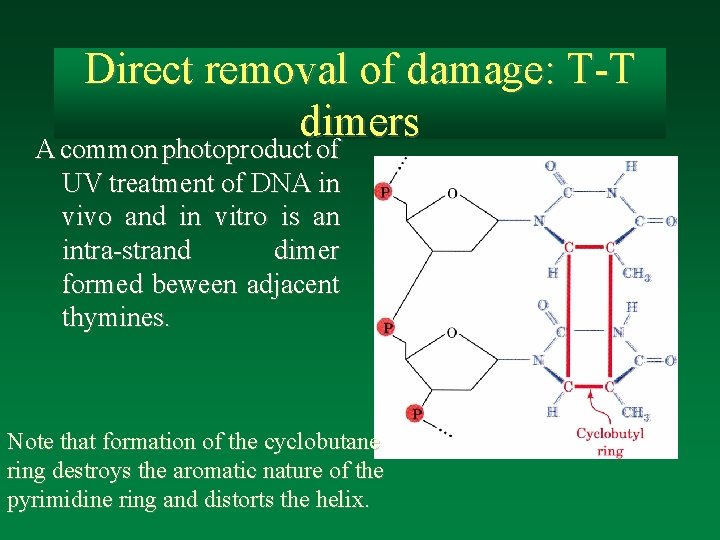
Direct removal of damage: T-T dimers A common photoproduct of UV treatment of DNA in vivo and in vitro is an intra-strand dimer formed beween adjacent thymines. Note that formation of the cyclobutane ring destroys the aromatic nature of the pyrimidine ring and distorts the helix.

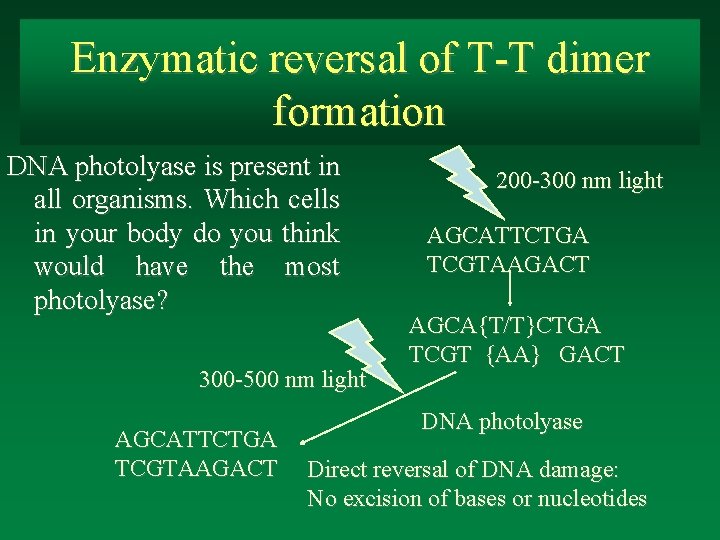
Enzymatic reversal of T-T dimer formation DNA photolyase is present in all organisms. Which cells in your body do you think would have the most photolyase? 300 -500 nm light AGCATTCTGA TCGTAAGACT 200 -300 nm light AGCATTCTGA TCGTAAGACT AGCA{T/T}CTGA TCGT {AA} GACT DNA photolyase Direct reversal of DNA damage: No excision of bases or nucleotides

Alkyltransferase detoxification of alkylated DNA by the Ada protein • A second type of direct reversal of DNA damage removes offending akyl groups from O 6 -alkyl guanine and methylated phosphate triesters. Alkylation of Cys 321 inactivates the protein-the protein “commits suicide”. Buried active site cysteine 321 covalently binds methyl group of O 6 m. G.

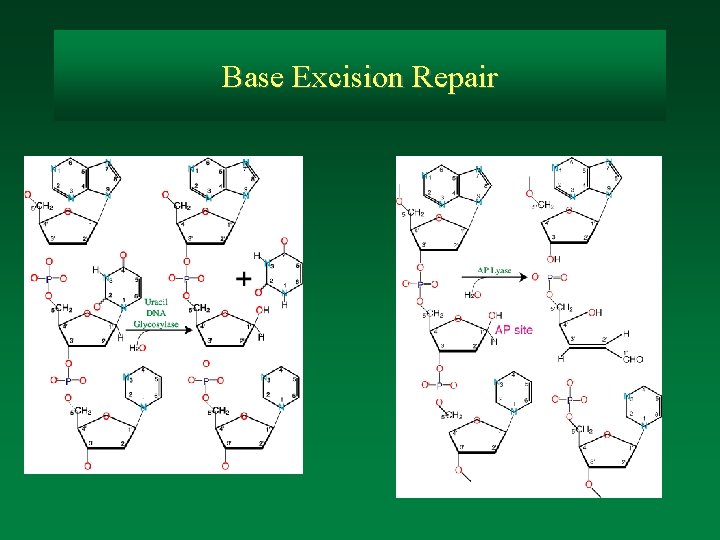
Base Excision Repair

Base Excision Repair

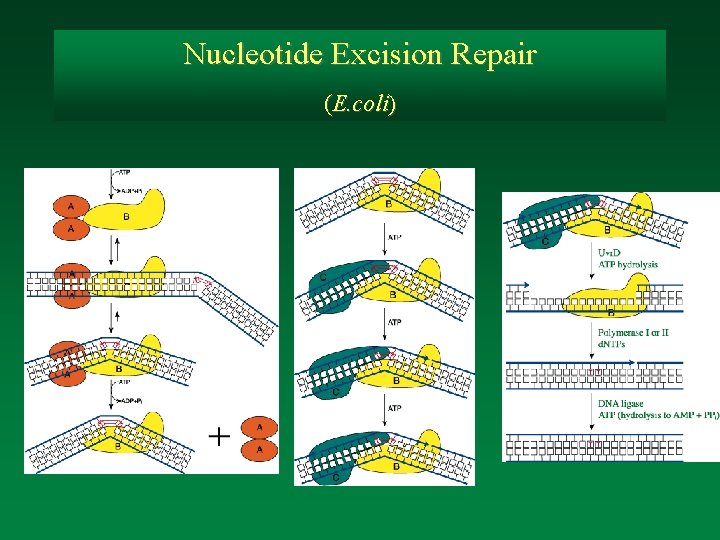
Nucleotide Excision Repair (E. coli)

Nucleotide Excision Repair (Global Genome Repair -Humans)

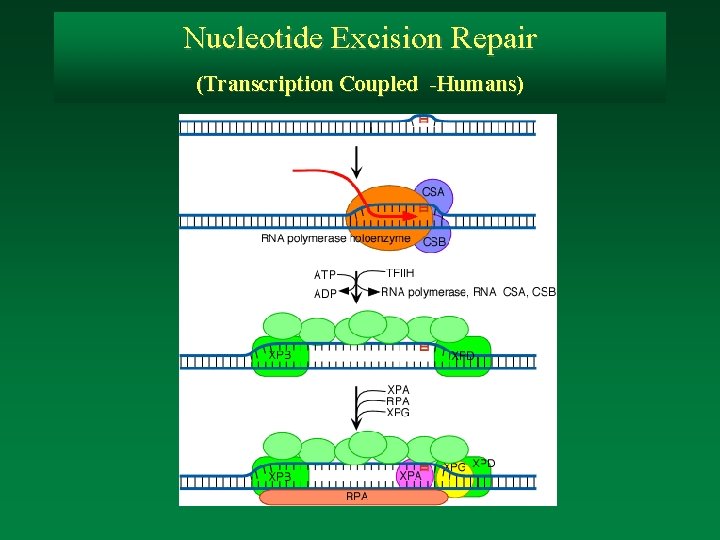
Nucleotide Excision Repair (Transcription Coupled -Humans)

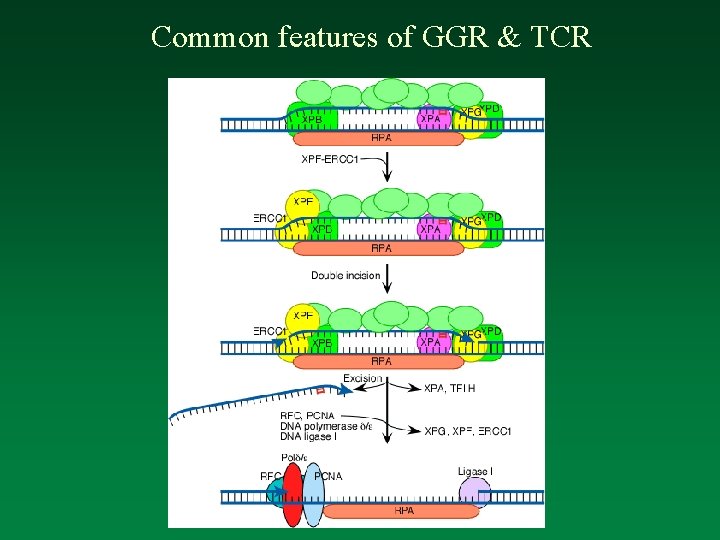
Common features of GGR & TCR

4. DNA glycosylases remove altered bases Deamination of cytosine, particularly 5 -methyl cytosine leaves uracil (thymine if 5 methyl cytosine) in the DNA. Uracil would pair as thymine during replication and thus cause a mutation. Uracil-N-glycosylase removes uracil from DNA. An endonuclease then cleaves the backbone at that site, creating a substrate for NER

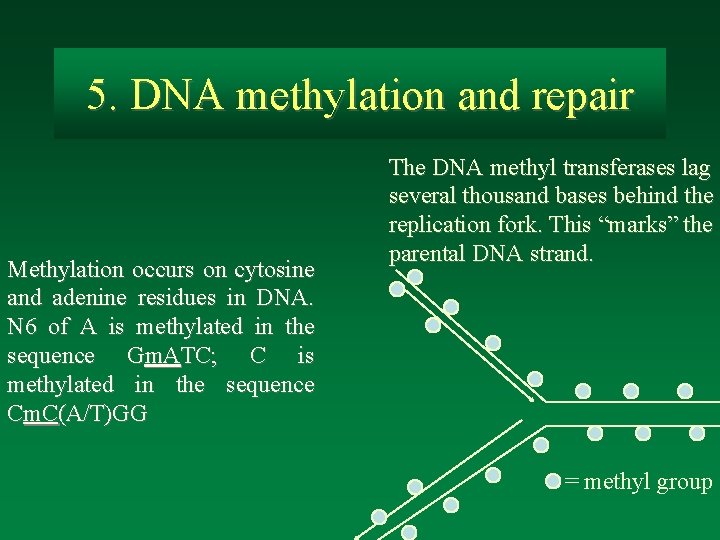
5. DNA methylation and repair Methylation occurs on cytosine and adenine residues in DNA. N 6 of A is methylated in the sequence Gm. ATC; C is methylated in the sequence Cm. C(A/T)GG The DNA methyl transferases lag several thousand bases behind the replication fork. This “marks” the parental DNA strand. = methyl group

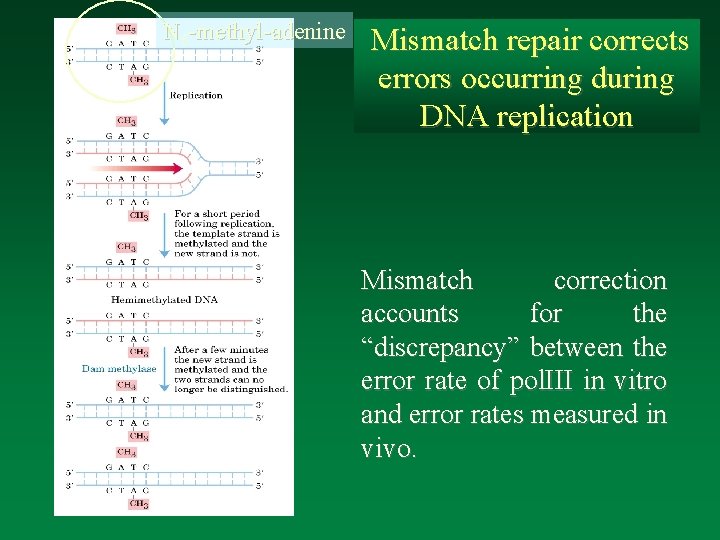
N 6 -methyl-adenine Mismatch repair corrects errors occurring during DNA replication Mismatch correction accounts for the “discrepancy” between the error rate of pol. III in vitro and error rates measured in vivo.

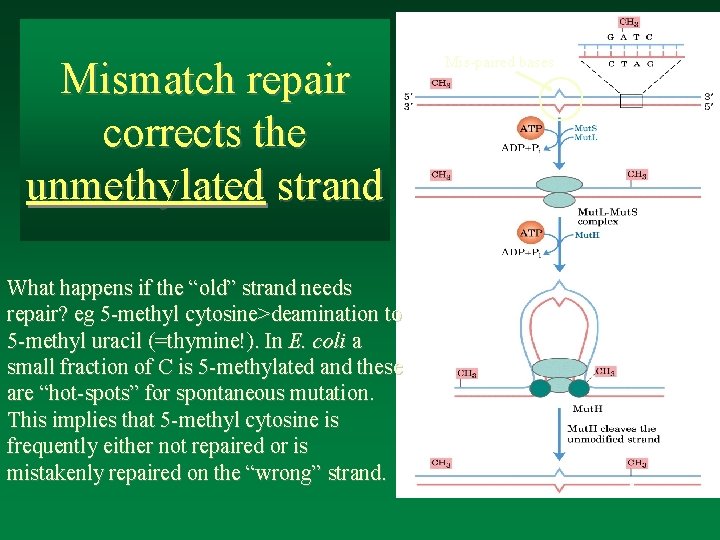
Mismatch repair corrects the unmethylated strand What happens if the “old” strand needs repair? eg 5 -methyl cytosine>deamination to 5 -methyl uracil (=thymine!). In E. coli a small fraction of C is 5 -methylated and these are “hot-spots” for spontaneous mutation. This implies that 5 -methyl cytosine is frequently either not repaired or is mistakenly repaired on the “wrong” strand. Mis-paired bases

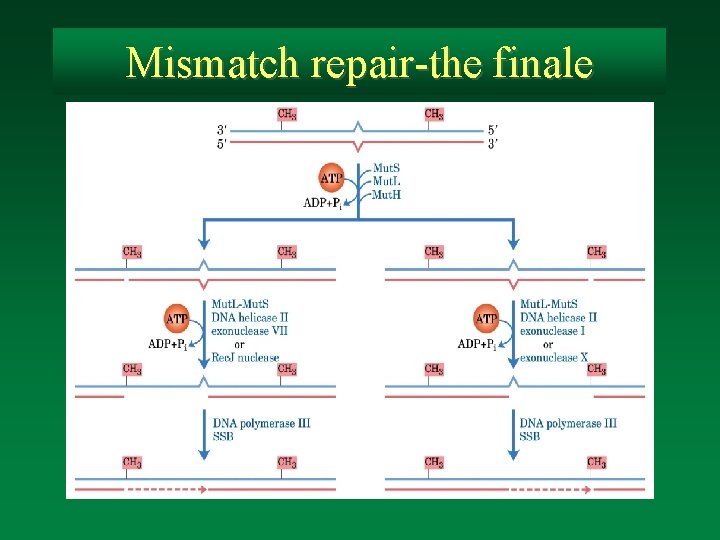
Mismatch repair-the finale

6. Error-prone or induced DNA repair- the SOS reponse UV’d T 4 phage UV UV’d T 4 phage E. coli Higher frequency of surviving phage, but many mutants. • Irradiation of bacteria before virus infection enhanced repair of damaged viral genes but led to mutations. This has an evolutionary advantage for the viral population since it increases the probability that some members will survive albeit in altered form Few surviving phage

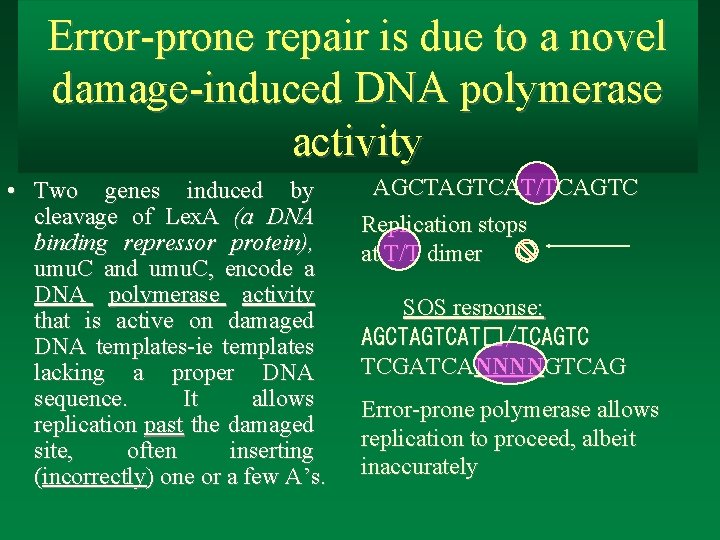
Error-prone repair is due to a novel damage-induced DNA polymerase activity • Two genes induced by cleavage of Lex. A (a DNA binding repressor protein), umu. C and umu. C, encode a DNA polymerase activity that is active on damaged DNA templates-ie templates lacking a proper DNA sequence. It allows replication past the damaged site, often inserting (incorrectly) one or a few A’s. AGCTAGTCAT/TCAGTC Replication stops at T/T dimer SOS response: AGCTAGTCAT�/TCAGTC TCGATCANNNNGTCAG Error-prone polymerase allows replication to proceed, albeit inaccurately

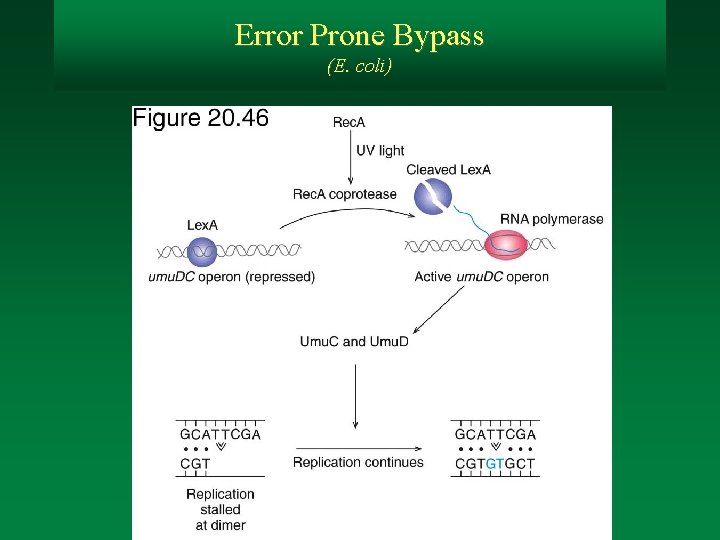
Error Prone Bypass (E. coli)

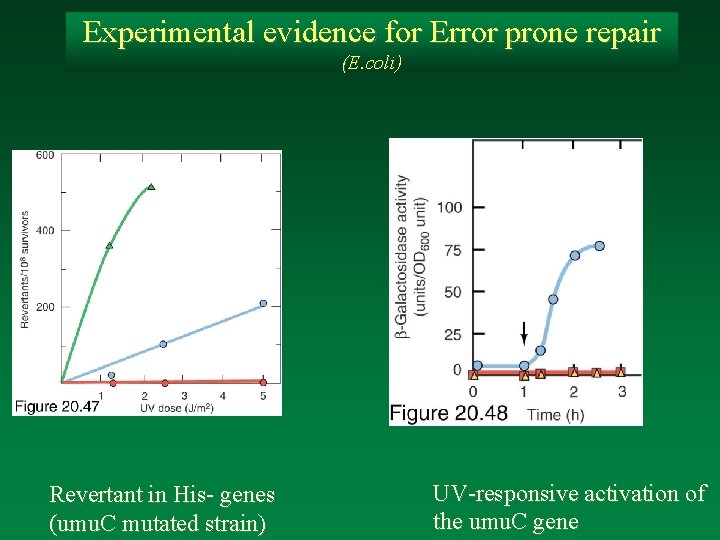
Experimental evidence for Error prone repair (E. coli) Revertant in His- genes (umu. C mutated strain) UV-responsive activation of the umu. C gene

Human mismatch repair genes and cancer • Yeast mutants defective in mismatch repair genes have unstable microsatellite sequences (repetitive tracts of mono- and dinucleotides). • Some human colon cancers also display microsatellite instability. • Are these due to defective mismatch repair genes? ANSWER: Yes: Genetic mapping of human nonpolyposis colon cancer genes identifies these genes as defective human mismatch repair genes. Can these be corrected? …. .

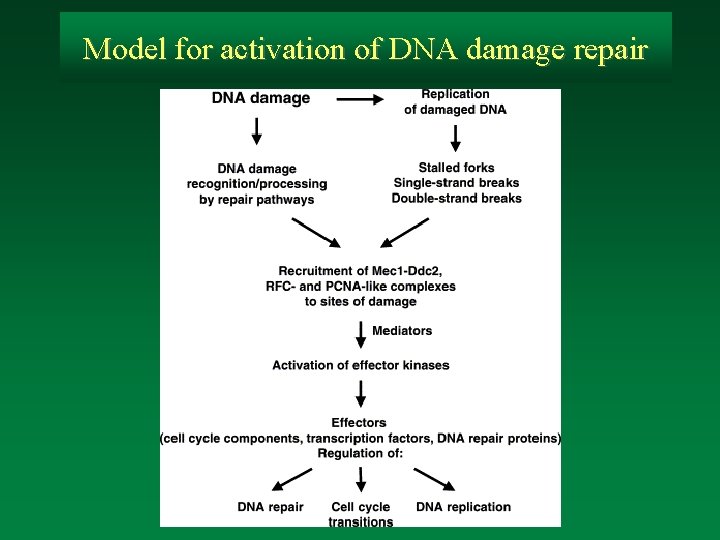
Model for activation of DNA damage repair

Damage & Repair • Multiple forms of DNA damage occur • These are repaired constantly by several mechanisms • Failure to repair damage leads to mutations • Often defects in damage sensing machinery or DNA repair processes can be correlated with increased incidence of diseases such as cancer

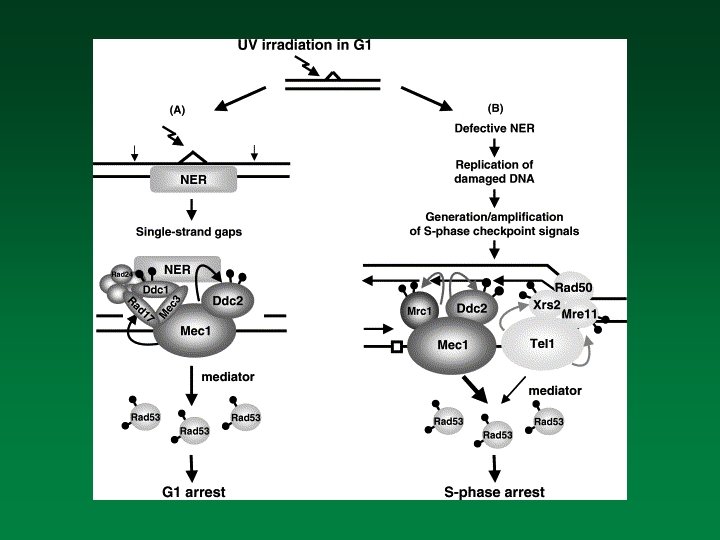
Factors involved in Damage Sensing




DNA Repair-summary • DNA repair mechanisms exist in all organisms to maintain the fidelity of the DNA sequence. • DNA is repaired during and after replication, and by constitutive and damage-inducible enzyme systems • Multiple repair mechanisms are necessary to correct errors arising during replication and to repair DNA damage by intrinsic and extrinsic agents. • Failure of DNA repair leads to mutations and cancer.
 Cuales son los tipos de mantenimiento correctivo
Cuales son los tipos de mantenimiento correctivo Taip samone pavercia mus bailiais
Taip samone pavercia mus bailiais Mutacin
Mutacin Depurination
Depurination Base excision repair
Base excision repair Hey bye bye
Hey bye bye Dna polymerase proofreading
Dna polymerase proofreading Dont ask why why why
Dont ask why why why Replication fork
Replication fork Bioflix activity dna replication dna replication diagram
Bioflix activity dna replication dna replication diagram Coding dna and non coding dna
Coding dna and non coding dna Replication process
Replication process Dna rna protein synthesis homework #2 dna replication
Dna rna protein synthesis homework #2 dna replication What is electrophoresis
What is electrophoresis Functions of nucleotides
Functions of nucleotides Why is dna important
Why is dna important Dna meaning
Dna meaning Why is dna replication considered semiconservative
Why is dna replication considered semiconservative Why dna is more stable than rna?
Why dna is more stable than rna? Tru count air clutch
Tru count air clutch Duct repair serpentine
Duct repair serpentine Decompressive laminectomy icd 10
Decompressive laminectomy icd 10 Ems equipment repair
Ems equipment repair Other initiated self repair example
Other initiated self repair example Control center centurylink
Control center centurylink Crack comparator card
Crack comparator card Shipbreaker equipment mount
Shipbreaker equipment mount Maag gearbox repair
Maag gearbox repair Verb 3 destroy
Verb 3 destroy Level of repair analysis template
Level of repair analysis template Bus garage repair dfd level 1
Bus garage repair dfd level 1 Indirect v direct hernia
Indirect v direct hernia Glencoe health chapter 6
Glencoe health chapter 6 Fix üçüncü hali
Fix üçüncü hali Hernia of linea alba
Hernia of linea alba Dfd level 0 and level 1
Dfd level 0 and level 1 Culvert repair techniques
Culvert repair techniques Power brake system
Power brake system Modified macpherson strut
Modified macpherson strut Chapter 42 gasoline injection diagnosis and repair
Chapter 42 gasoline injection diagnosis and repair Heavy duty truck axle repair
Heavy duty truck axle repair Write it right bureau of automotive repair
Write it right bureau of automotive repair Chapter 44 automotive wiring and wire repair
Chapter 44 automotive wiring and wire repair Pcb replacement training
Pcb replacement training Wells fargo identity theft repair kit
Wells fargo identity theft repair kit Samsung pba repair
Samsung pba repair